Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200447 |
| Name | oriT_ICEMlSymSU343-α |
| Organism | Mesorhizobium loti strain SU343 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP033368 (94281..94341 [+], 61 nt) |
| oriT length | 61 nt |
| IRs (inverted repeats) | 26..33, 37..44 (ACCGCCTC..GAGGCGGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 61 nt
>oriT_ICEMlSymSU343-α
AAAACGATGTGCAGACAAAGACTTGACCGCCTCGTGGAGGCGGTGCCTTTATCTTGCCTTA
AAAACGATGTGCAGACAAAGACTTGACCGCCTCGTGGAGGCGGTGCCTTTATCTTGCCTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file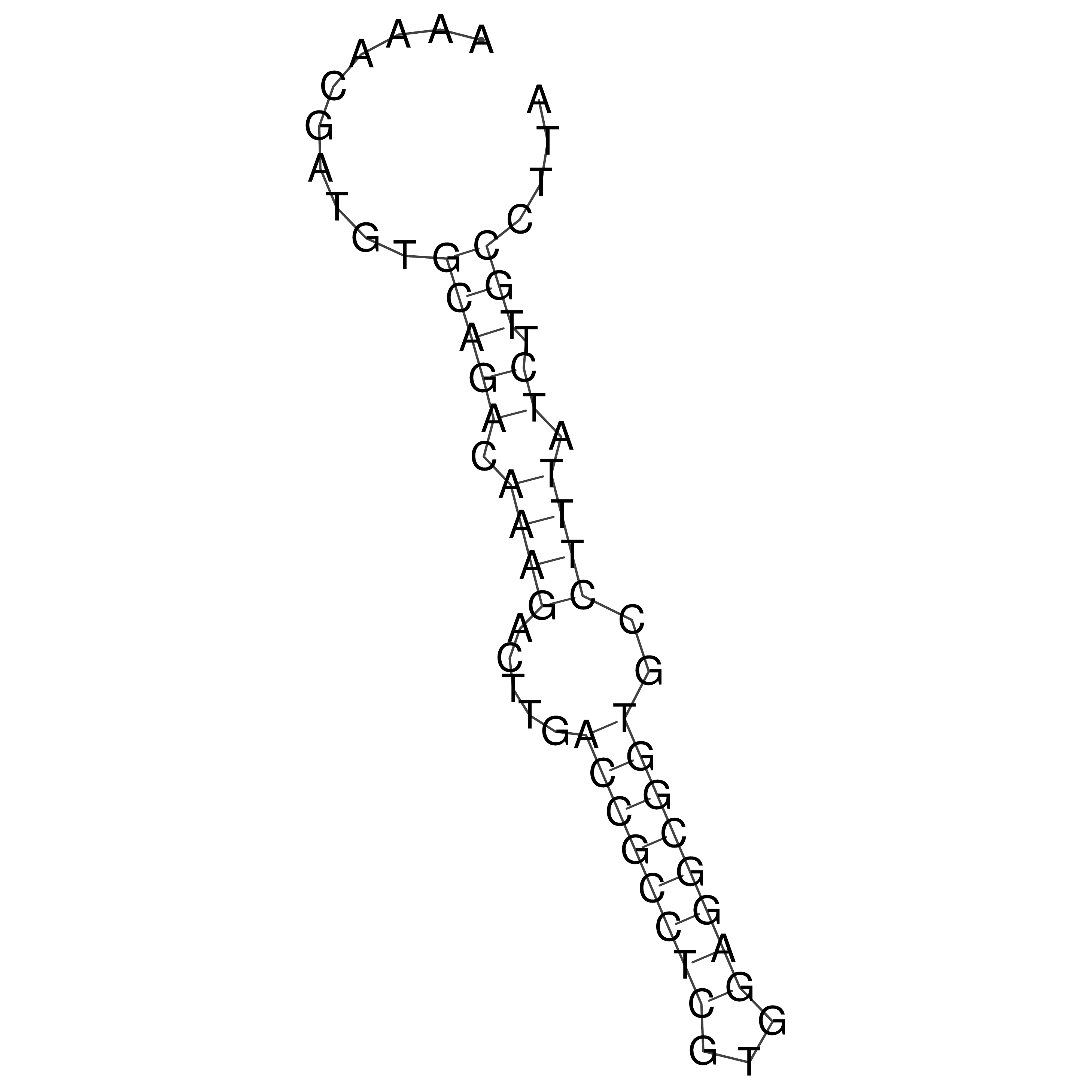
T4CP
| ID | 17049 | GenBank | QKD13264 |
| Name | t4cp2_EFV37_31315_ICEMlSymSU343-α |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 90140.67 Da Isoelectric Point: 5.9573
>QKD13264.1 conjugal transfer protein TrbE [Mesorhizobium loti]
MLNLSEYRGKADRLADHLPWAALVAPGIVLNKDGSVQRTLRFRGPDLESATEAELIGICARANNALRRLG
SGWTLFFEAERIEALGYPSSRFPDAASWLVDEERRAAFEGKVAHFESRYHLTLLSMPPPDAQARAESALV
DSHHSEGERDWRQEIARFRDETDRVLDLLSGFMPEVRLLDDAETLTYLHDTISTRRHPVAVPETPMYLDG
ILVDVPLTGGLEPMLGEQHLRTLTILGFPNLTRPGILDALNHQDFAYRWMTRFIPLGKTEATKTLTRLRR
QWFAKRKSMVAILREVVSNEPVPLVDSDADNKALDADEALQALGGDHIGFGYLTATVTVWDEDRQAAAEK
LRAVERIINGLGFTTIRESVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAEI
TGREAPPLLIAETSGSTPFRLSSHVEDVGHMLIVGPTGAGKSVLLALIALQFRRYAGAQVYVFDKGNSAR
AATLAMGGEHHVLGSDGALAFQPLRNIGDQGRRSWAAEWIAGLIAHENVALTPEVKEAIWSALNSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHSQSAVLPVLTYLF
HRLEERFDGRPTLLMLDEAWVDLDNPLFAARIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQARTAYERFGLNNRQIELIARATPKRHYYLQSRCGNRRFELGLGPIALALCGASGPA
AQSLIDRVLSEHGQDSFVFRFLGACGLDWAAELLQQFPQPTKEQSS
MLNLSEYRGKADRLADHLPWAALVAPGIVLNKDGSVQRTLRFRGPDLESATEAELIGICARANNALRRLG
SGWTLFFEAERIEALGYPSSRFPDAASWLVDEERRAAFEGKVAHFESRYHLTLLSMPPPDAQARAESALV
DSHHSEGERDWRQEIARFRDETDRVLDLLSGFMPEVRLLDDAETLTYLHDTISTRRHPVAVPETPMYLDG
ILVDVPLTGGLEPMLGEQHLRTLTILGFPNLTRPGILDALNHQDFAYRWMTRFIPLGKTEATKTLTRLRR
QWFAKRKSMVAILREVVSNEPVPLVDSDADNKALDADEALQALGGDHIGFGYLTATVTVWDEDRQAAAEK
LRAVERIINGLGFTTIRESVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAEI
TGREAPPLLIAETSGSTPFRLSSHVEDVGHMLIVGPTGAGKSVLLALIALQFRRYAGAQVYVFDKGNSAR
AATLAMGGEHHVLGSDGALAFQPLRNIGDQGRRSWAAEWIAGLIAHENVALTPEVKEAIWSALNSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHSQSAVLPVLTYLF
HRLEERFDGRPTLLMLDEAWVDLDNPLFAARIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQARTAYERFGLNNRQIELIARATPKRHYYLQSRCGNRRFELGLGPIALALCGASGPA
AQSLIDRVLSEHGQDSFVFRFLGACGLDWAAELLQQFPQPTKEQSS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17050 | GenBank | QKD12168 |
| Name | t4cp2_EFV37_31340_ICEMlSymSU343-α |
UniProt ID | _ |
| Length | 676 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 676 a.a. Molecular weight: 75073.54 Da Isoelectric Point: 6.6518
>QKD12168.1 conjugal transfer protein TraG [Mesorhizobium loti]
MTPTKLLIGQIAIVFAIVLLGVWAATQWCAHMLAFQEQLGAPWFVAAGWPIYEPWKLLEWWFQFDAYAPE
VFDKAGMLAGASGFMGCAAAIAGSLWRARQRGLVTTYGSSRWAVTREIEKAGLFQPAGVFLGKLKDRYLR
HDGPEHVMAFAPTRSGKGVGLVVPTLLSWSGSAVIHDIKGENWQLSSGWRSRFSHCLLFNPTDPRSARYN
PLLEVRKGSDEIRDVQNIADILVDPEGALERRNHWEKTSHSLLVGAILHVLYAEEEKTLARVATFLSDPQ
RSFAATLRRMMTTNHLGAANKPQVHPVVASAAREVLNKSENERSGVLSTAMSFLGLYRDPTVAAVTSDCD
WRIADLMDAEWPMSLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPKKSRKHQLLMMLDEFPALGR
LDFFETALAFMAGYGIRAYLIAQSLNQISKAYGDNNAILDNCHLRIAFSSNDERTAKRISDALGTATELR
SMRNYAGHRLAPWLSHVMVSRQETARPLLTPGEVMQLPPADELVLVSGLPPIRAKKLRYYEDHNFTERVL
PSPVLRDGPYADCPMSRPDDWTGQVRGVDRRLASDDESAGAALADEGGVQQQRHPGLPEEHTAVTQEPDQ
DDLSRPADDDSEAMADKRVMDRISTVAGAYGVNEGRGDDKDIVPGF
MTPTKLLIGQIAIVFAIVLLGVWAATQWCAHMLAFQEQLGAPWFVAAGWPIYEPWKLLEWWFQFDAYAPE
VFDKAGMLAGASGFMGCAAAIAGSLWRARQRGLVTTYGSSRWAVTREIEKAGLFQPAGVFLGKLKDRYLR
HDGPEHVMAFAPTRSGKGVGLVVPTLLSWSGSAVIHDIKGENWQLSSGWRSRFSHCLLFNPTDPRSARYN
PLLEVRKGSDEIRDVQNIADILVDPEGALERRNHWEKTSHSLLVGAILHVLYAEEEKTLARVATFLSDPQ
RSFAATLRRMMTTNHLGAANKPQVHPVVASAAREVLNKSENERSGVLSTAMSFLGLYRDPTVAAVTSDCD
WRIADLMDAEWPMSLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPKKSRKHQLLMMLDEFPALGR
LDFFETALAFMAGYGIRAYLIAQSLNQISKAYGDNNAILDNCHLRIAFSSNDERTAKRISDALGTATELR
SMRNYAGHRLAPWLSHVMVSRQETARPLLTPGEVMQLPPADELVLVSGLPPIRAKKLRYYEDHNFTERVL
PSPVLRDGPYADCPMSRPDDWTGQVRGVDRRLASDDESAGAALADEGGVQQQRHPGLPEEHTAVTQEPDQ
DDLSRPADDDSEAMADKRVMDRISTVAGAYGVNEGRGDDKDIVPGF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17051 | GenBank | QKD12195 |
| Name | t4cp2_EFV37_31525_ICEMlSymSU343-α |
UniProt ID | _ |
| Length | 563 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 563 a.a. Molecular weight: 62321.66 Da Isoelectric Point: 8.3286
>QKD12195.1 type IV secretion system protein VirD4 [Mesorhizobium loti]
MSSTKTTPPNIALSIACSLALGFCAASLYATFRHGGSGEALMTFDVRAFWFETPFYLGFATPVFYRGIAI
VLATSAVVLLIQQVVLRRKLEHHGTARWARVDEMRCTGYLRRYRSVTGPVFGKTSGPFWPGYYLTNGEQP
HSLIVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKATGDNVFKFAPLDSERRTNCYNPL
LDLIALPPQLQFTEARRLAANLITAKGEGAEGFVSGARDIFVAGILACIERGTPTIGAVYDLFAQPGEKF
KLFAQLAMETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPAVKAATSRSDFSVYDLRRRRTC
IYLCVGPNDLEVIAPLIRLFFQQIVSILQRSLPRQDEKNEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMLIIQSLSALTGAYGEAGKQNFLSNTGLQVFMATADDETPTYISKAIGEYTFEAKSISYSQARTFDWNI
QNSHQGAPLLRPEQVRLLDEDYEIVLIKGLPPLQLRKVRYYSDRILKRIFESQTGALPEPAPLMEAHPDD
TDG
MSSTKTTPPNIALSIACSLALGFCAASLYATFRHGGSGEALMTFDVRAFWFETPFYLGFATPVFYRGIAI
VLATSAVVLLIQQVVLRRKLEHHGTARWARVDEMRCTGYLRRYRSVTGPVFGKTSGPFWPGYYLTNGEQP
HSLIVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKATGDNVFKFAPLDSERRTNCYNPL
LDLIALPPQLQFTEARRLAANLITAKGEGAEGFVSGARDIFVAGILACIERGTPTIGAVYDLFAQPGEKF
KLFAQLAMETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPAVKAATSRSDFSVYDLRRRRTC
IYLCVGPNDLEVIAPLIRLFFQQIVSILQRSLPRQDEKNEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMLIIQSLSALTGAYGEAGKQNFLSNTGLQVFMATADDETPTYISKAIGEYTFEAKSISYSQARTFDWNI
QNSHQGAPLLRPEQVRLLDEDYEIVLIKGLPPLQLRKVRYYSDRILKRIFESQTGALPEPAPLMEAHPDD
TDG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 6295965..6305022
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EFV37_31255 | 6291021..6291170 | - | 150 | QKD12153 | CcoQ/FixQ family Cbb3-type cytochrome c oxidase assembly chaperone | - |
| EFV37_31260 | 6291181..6291912 | - | 732 | QKD12154 | cytochrome-c oxidase, cbb3-type subunit II | - |
| EFV37_31265 | 6291924..6293534 | - | 1611 | QKD12155 | cytochrome-c oxidase, cbb3-type subunit I | - |
| EFV37_31270 | 6293664..6294203 | + | 540 | QKD12156 | hemerythrin domain-containing protein | - |
| EFV37_31275 | 6294207..6294962 | - | 756 | QKD12157 | Crp/Fnr family transcriptional regulator | - |
| EFV37_31280 | 6295107..6295631 | + | 525 | Protein_6194 | radical SAM protein | - |
| EFV37_31285 | 6295707..6295952 | - | 246 | QKD12158 | DUF2274 domain-containing protein | - |
| EFV37_31290 | 6295965..6297179 | - | 1215 | QKD12159 | TrbI/VirB10 family protein | virB10 |
| EFV37_31295 | 6297191..6298219 | - | 1029 | QKD12160 | P-type conjugative transfer protein TrbG | virB9 |
| EFV37_31300 | 6298216..6298941 | - | 726 | QKD12161 | conjugal transfer protein TrbF | virB8 |
| EFV37_31305 | 6298942..6300264 | - | 1323 | QKD12162 | P-type conjugative transfer protein TrbL | virB6 |
| EFV37_31310 | 6300296..6301021 | - | 726 | QKD12163 | P-type conjugative transfer protein TrbJ | virB5 |
| EFV37_31315 | 6301018..6303468 | - | 2451 | QKD13264 | conjugal transfer protein TrbE | virb4 |
| EFV37_31320 | 6303462..6303734 | - | 273 | QKD12164 | conjugal transfer protein | virB3 |
| EFV37_31325 | 6303734..6304042 | - | 309 | QKD12165 | conjugal transfer protein TrbC | virB2 |
| EFV37_31330 | 6304054..6305022 | - | 969 | QKD12166 | P-type conjugative transfer ATPase TrbB | virB11 |
| EFV37_31335 | 6305019..6305432 | - | 414 | QKD12167 | CopG family transcriptional regulator | - |
| EFV37_31340 | 6305551..6307581 | - | 2031 | QKD12168 | conjugal transfer protein TraG | - |
| EFV37_31345 | 6307711..6307948 | + | 238 | Protein_6207 | XRE family transcriptional regulator | - |
| EFV37_31350 | 6308246..6308635 | - | 390 | QKD12169 | hypothetical protein | - |
| EFV37_31355 | 6308626..6309033 | - | 408 | QKD12170 | hypothetical protein | - |
| EFV37_31360 | 6309328..6309618 | + | 291 | QKD12171 | hypothetical protein | - |
Region 2: 6325709..6335131
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EFV37_31410 | 6321340..6321470 | + | 131 | Protein_6220 | IS630 family transposase | - |
| EFV37_31415 | 6321470..6322082 | + | 613 | Protein_6221 | ISKra4 family transposase | - |
| EFV37_31420 | 6323055..6325595 | + | 2541 | QKD12179 | two-component system VirA-like sensor kinase | - |
| EFV37_31425 | 6325709..6326749 | - | 1041 | QKD12180 | P-type DNA transfer ATPase VirB11 | virB11 |
| EFV37_31430 | 6326733..6327872 | - | 1140 | QKD12181 | type IV secretion system protein VirB10 | virB10 |
| EFV37_31435 | 6327869..6328750 | - | 882 | QKD12182 | type IV secretion system protein VirB9 | virB9 |
| EFV37_31440 | 6328747..6329460 | - | 714 | QKD12183 | type IV secretion system protein VirB8 | virB8 |
| EFV37_31445 | 6329457..6329615 | - | 159 | QKD12184 | type IV secretion system lipoprotein VirB7 | - |
| EFV37_31450 | 6329642..6330526 | - | 885 | QKD12185 | type IV secretion system protein | virB6 |
| EFV37_31455 | 6330585..6331265 | - | 681 | QKD12186 | type IV secretion system protein VirB5 | - |
| EFV37_31460 | 6331279..6333648 | - | 2370 | QKD12187 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| EFV37_31465 | 6333648..6333974 | - | 327 | QKD12188 | type IV secretion system protein VirB3 | virB3 |
| EFV37_31470 | 6333974..6334339 | - | 366 | QKD12189 | pilin major subunit VirB2 | virB2 |
| EFV37_31475 | 6334355..6335131 | - | 777 | QKD12190 | type IV secretion system lytic transglycosylase VirB1 | virB1 |
| EFV37_31480 | 6335219..6335416 | - | 198 | QKD12191 | hypothetical protein | - |
| EFV37_31485 | 6336195..6336914 | + | 720 | QKD12192 | response regulator | - |
| EFV37_31490 | 6336883..6337161 | + | 279 | QKD12193 | hypothetical protein | - |
| EFV37_31495 | 6337211..6337429 | - | 219 | QKD12194 | hypothetical protein | - |
Host bacterium
| ID | 398 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEMlSymSU343-α | GenBank | CP033368 |
| Element size | 6934480 bp | Coordinate of oriT [Strand] | 94281..94341 [+] |
| Host bacterium | Mesorhizobium loti strain SU343 | Coordinate of element | 6285206..6807512 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixS, fixI, nifX, fixG, fixP, fixQ, fixO, fixN, nodM, nodD, nolL, nodB, a9174_31165, nodJ, nodI, nodC, nodA, nodS, mlr6097, nifU, nifN, nifE, nifK, nifD, nifH, nifQ, mLTONO_5203, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, fixT, nolK, noeL, nodZ, nodE, nodF, nodU |
| Anti-CRISPR | AcrIIC1 |