Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200443 |
| Name | oriT_ICEMsp.SymNZP2298 |
| Organism | Mesorhizobium sp. NZP2298 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP033365 (146676..146735 [+], 60 nt) |
| oriT length | 60 nt |
| IRs (inverted repeats) | 26..32, 36..42 (CCGCCTC..GAGGCGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 60 nt
>oriT_ICEMsp.SymNZP2298
AAACGATGTGCAGACAAAGACTTCGCCGCCTCATGGAGGCGGTGCCTTTATCTTGCCTTA
AAACGATGTGCAGACAAAGACTTCGCCGCCTCATGGAGGCGGTGCCTTTATCTTGCCTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file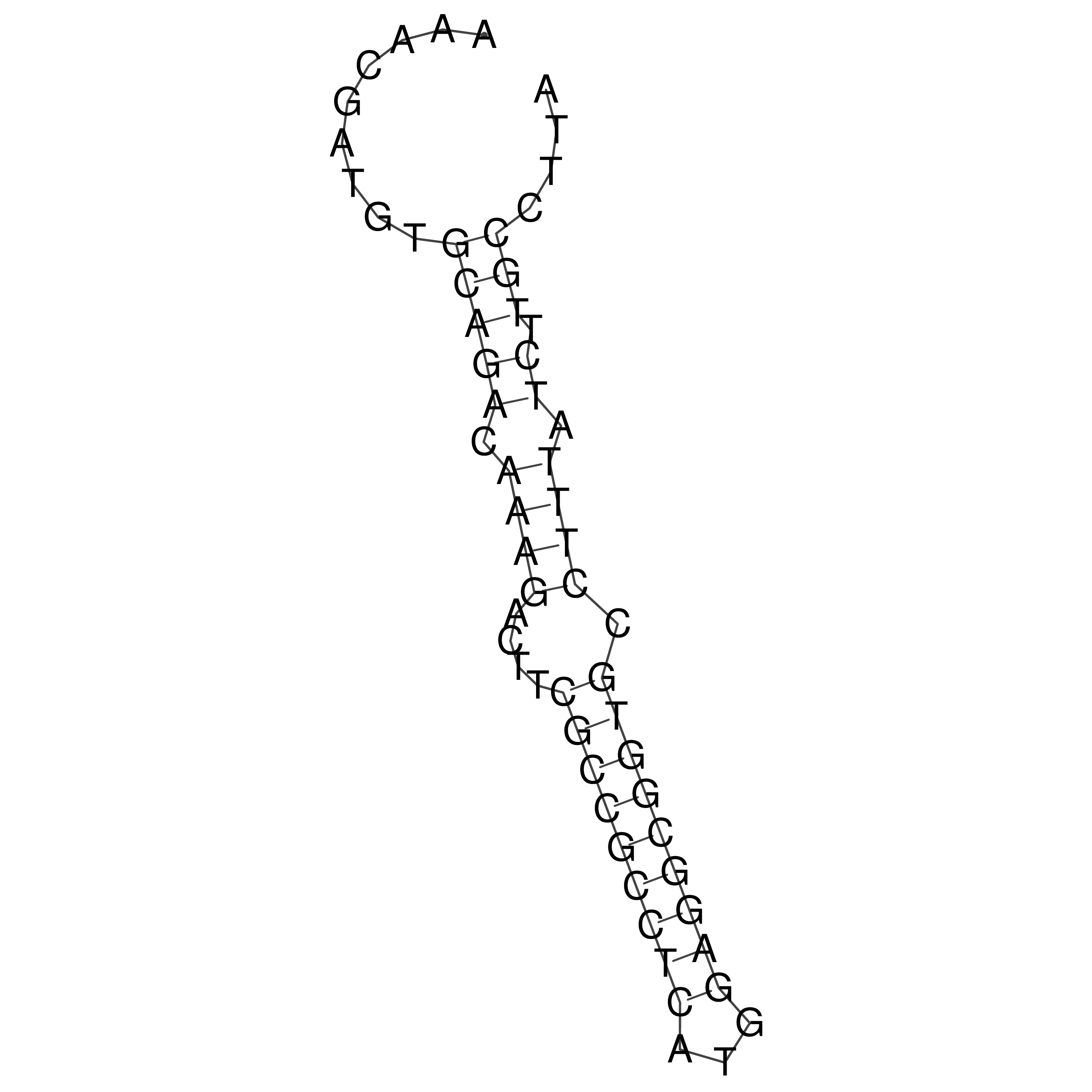
T4CP
| ID | 17038 | GenBank | QKC98722 |
| Name | t4cp2_EB231_31845_ICEMsp.SymNZP2298 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 90285.86 Da Isoelectric Point: 6.1178
>QKC98722.1 conjugal transfer protein TrbE [Mesorhizobium sp. NZP2298]
MLNLSEYRGKADRLADHLPWAALVAPGIVLNKDGSVQRTIRFRGPDLESSTEAELIGICARANNALRRLG
SGWTLFFEAERIEALGYPSSRFPDAASWLVDEERRAAFEGKVAHFESRYHLTLLSMPPPDAQARAESALV
DSHHSEGERDWRQEMARFRDETDRVLDLLSGFMPEVRLLDDAETLTYLHDTISTRRHPVAVPETPMYLDG
ILVDVPLTGGLEPMLGEQHLRTLTILGFPNLTRPGILDALNHQDFAYRWMTRFIPLGKTEATKTLTRLRR
QWFAKRKSIVAILREVVSNEPVPLVDSDADNKALDADEALQALGGDHVGFGYLTATVTVWDEDRQAAAEK
LRAVERIINGLGFTTIRESVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAEV
TGREAPPLLIAETSGSTPFRLSSHVEDVGHMLIVGPTGAGKSVLLALIALQFRRYAGAQVYVFDKGNSAR
AATLAMGGEHHVLGSDGALAFQPLRNIGDQGRRSWAAEWIAGLIAHENVTLTPEVKEAIWSALNSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHRQSAVLPVLTYLF
HRLEERFDGRPTLLMLDEAWVYLDNPLFAARIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQARTAYERFGLNNRQIELIARATPKRHYYLQSRCGNRLFELGLGPIALALCGASGPA
AQSLIDRVLSEHGQDSFVFRFLGARGLDWAAELLQQFPQPTKEQSS
MLNLSEYRGKADRLADHLPWAALVAPGIVLNKDGSVQRTIRFRGPDLESSTEAELIGICARANNALRRLG
SGWTLFFEAERIEALGYPSSRFPDAASWLVDEERRAAFEGKVAHFESRYHLTLLSMPPPDAQARAESALV
DSHHSEGERDWRQEMARFRDETDRVLDLLSGFMPEVRLLDDAETLTYLHDTISTRRHPVAVPETPMYLDG
ILVDVPLTGGLEPMLGEQHLRTLTILGFPNLTRPGILDALNHQDFAYRWMTRFIPLGKTEATKTLTRLRR
QWFAKRKSIVAILREVVSNEPVPLVDSDADNKALDADEALQALGGDHVGFGYLTATVTVWDEDRQAAAEK
LRAVERIINGLGFTTIRESVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAEV
TGREAPPLLIAETSGSTPFRLSSHVEDVGHMLIVGPTGAGKSVLLALIALQFRRYAGAQVYVFDKGNSAR
AATLAMGGEHHVLGSDGALAFQPLRNIGDQGRRSWAAEWIAGLIAHENVTLTPEVKEAIWSALNSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHRQSAVLPVLTYLF
HRLEERFDGRPTLLMLDEAWVYLDNPLFAARIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQARTAYERFGLNNRQIELIARATPKRHYYLQSRCGNRLFELGLGPIALALCGASGPA
AQSLIDRVLSEHGQDSFVFRFLGARGLDWAAELLQQFPQPTKEQSS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17039 | GenBank | QKC98727 |
| Name | t4cp2_EB231_31870_ICEMsp.SymNZP2298 |
UniProt ID | _ |
| Length | 676 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 676 a.a. Molecular weight: 75105.54 Da Isoelectric Point: 6.6518
>QKC98727.1 conjugal transfer protein TraG [Mesorhizobium sp. NZP2298]
MTPTKLLIGQIAIVFAIVLLGVWAATQWCAHMLAFQEQLGAPWFVAAGWPIYEPWKLLEWWFQFDAYAPE
VFDKAGMLAGTSGFMGCAAAIAGSLWRARQRGLVTTYGSSRWAATREIEKAGLFQPAGVFLGKLKDRYLR
HDGPEHVMAFAPTRSGKGVGLVVPTLLSWSGSAVIHDIKGENWQLSSGWRSRFSHCLLFNPTDPRSARYN
PLLEVRKGSDEIRDVQNIADILVDPEGALERRNHWEKTSHSLLVGAILHVLYAEEEKTLARVATFLSDPQ
RSFAATLRRMMTTNHLGSANKPQVHPVVASAAREVLNKSENERSGVLSTAMSFLGLYRDPTVAAVTSDCD
WRIADLMDAEWPMSLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPKKSRKHQLLMMLDEFPALGR
LDFFETALAFMAGYGIRAYLIAQSLNQISKAYGDNNAILDNCHLRIAFSSNDERTAKRISDALGTATELR
SMRNYAGHRLAPWLSHVMVSRQETARPLLTPGEVMQLPPADELVLVSGLPPIRAKKLRYYEDHNFTERVL
PSPVLRDGPYADCPMSRPDDWTGQVRGVDRRLASDDESAGAALADEGGVQQQRHPGLPEEHTAVTQEPDQ
DDLSRPADDDSEAMADKRVMDRISTVAGAYGINEGRGDDKDIVPGF
MTPTKLLIGQIAIVFAIVLLGVWAATQWCAHMLAFQEQLGAPWFVAAGWPIYEPWKLLEWWFQFDAYAPE
VFDKAGMLAGTSGFMGCAAAIAGSLWRARQRGLVTTYGSSRWAATREIEKAGLFQPAGVFLGKLKDRYLR
HDGPEHVMAFAPTRSGKGVGLVVPTLLSWSGSAVIHDIKGENWQLSSGWRSRFSHCLLFNPTDPRSARYN
PLLEVRKGSDEIRDVQNIADILVDPEGALERRNHWEKTSHSLLVGAILHVLYAEEEKTLARVATFLSDPQ
RSFAATLRRMMTTNHLGSANKPQVHPVVASAAREVLNKSENERSGVLSTAMSFLGLYRDPTVAAVTSDCD
WRIADLMDAEWPMSLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPKKSRKHQLLMMLDEFPALGR
LDFFETALAFMAGYGIRAYLIAQSLNQISKAYGDNNAILDNCHLRIAFSSNDERTAKRISDALGTATELR
SMRNYAGHRLAPWLSHVMVSRQETARPLLTPGEVMQLPPADELVLVSGLPPIRAKKLRYYEDHNFTERVL
PSPVLRDGPYADCPMSRPDDWTGQVRGVDRRLASDDESAGAALADEGGVQQQRHPGLPEEHTAVTQEPDQ
DDLSRPADDDSEAMADKRVMDRISTVAGAYGINEGRGDDKDIVPGF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17040 | GenBank | QKC98751 |
| Name | t4cp2_EB231_32050_ICEMsp.SymNZP2298 |
UniProt ID | _ |
| Length | 563 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 563 a.a. Molecular weight: 62345.60 Da Isoelectric Point: 8.7905
>QKC98751.1 type IV secretion system protein VirD4 [Mesorhizobium sp. NZP2298]
MSSTKTTSPNIAISIACSLALGFCAASLYATFRHGGSGEALMAFDVRAFWFETPFYLGFATPVFYQGAAI
VLSTSAVVLLIQQFVLRRKLEHHGTARWARVDEMRRTGYLRRYRGVTGPVFGKTSGPFWPGYYLTNGEQP
HSLIVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKAAGHEVFKFAPLDSERRTNCYNPL
LDLIALHPQQQFTEARRLAANLITAKGEGAEGFVSGARDIFVAGILACIERGTPTIGAVYDLFAQPGEKF
KLFAQLAMETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPAVKAATSRSDFSVYDLRRRRTC
IYLCVGPNDLEVIAPLIRLFFQQIVSILQRSLPRQDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMLIIQSLSALTGAYGEAGKQNFLSNTGLQVFMATADDETPNYISKAIGEYTFEAKSISYSQSRAFDRNI
QNSFQGAPLLRPEQVRLLNDEYEIVLIKGLPPLQLRKVRYFSDRILKRIFESQTGALPEPAPLMEAHPDD
TDG
MSSTKTTSPNIAISIACSLALGFCAASLYATFRHGGSGEALMAFDVRAFWFETPFYLGFATPVFYQGAAI
VLSTSAVVLLIQQFVLRRKLEHHGTARWARVDEMRRTGYLRRYRGVTGPVFGKTSGPFWPGYYLTNGEQP
HSLIVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKAAGHEVFKFAPLDSERRTNCYNPL
LDLIALHPQQQFTEARRLAANLITAKGEGAEGFVSGARDIFVAGILACIERGTPTIGAVYDLFAQPGEKF
KLFAQLAMETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPAVKAATSRSDFSVYDLRRRRTC
IYLCVGPNDLEVIAPLIRLFFQQIVSILQRSLPRQDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMLIIQSLSALTGAYGEAGKQNFLSNTGLQVFMATADDETPNYISKAIGEYTFEAKSISYSQSRAFDRNI
QNSFQGAPLLRPEQVRLLNDEYEIVLIKGLPPLQLRKVRYFSDRILKRIFESQTGALPEPAPLMEAHPDD
TDG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 6626037..6635094
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EB231_31800 | 6622411..6622950 | + | 540 | QKC98714 | hemerythrin domain-containing protein | - |
| EB231_31805 | 6622947..6623711 | - | 765 | QKC98715 | Crp/Fnr family transcriptional regulator | - |
| EB231_31810 | 6623869..6625218 | + | 1350 | QKC98716 | oxygen-independent coproporphyrinogen III oxidase | - |
| EB231_31815 | 6625755..6626024 | - | 270 | QKC98717 | DUF2274 domain-containing protein | - |
| EB231_31820 | 6626037..6627251 | - | 1215 | QKC99971 | TrbI/VirB10 family protein | virB10 |
| EB231_31825 | 6627263..6628291 | - | 1029 | QKC98718 | P-type conjugative transfer protein TrbG | virB9 |
| EB231_31830 | 6628288..6629013 | - | 726 | QKC98719 | conjugal transfer protein TrbF | virB8 |
| EB231_31835 | 6629014..6630336 | - | 1323 | QKC98720 | P-type conjugative transfer protein TrbL | virB6 |
| EB231_31840 | 6630368..6631093 | - | 726 | QKC98721 | P-type conjugative transfer protein TrbJ | virB5 |
| EB231_31845 | 6631090..6633540 | - | 2451 | QKC98722 | conjugal transfer protein TrbE | virb4 |
| EB231_31850 | 6633534..6633806 | - | 273 | QKC98723 | conjugal transfer protein | virB3 |
| EB231_31855 | 6633806..6634114 | - | 309 | QKC98724 | conjugal transfer protein TrbC | virB2 |
| EB231_31860 | 6634126..6635094 | - | 969 | QKC98725 | P-type conjugative transfer ATPase TrbB | virB11 |
| EB231_31865 | 6635091..6635504 | - | 414 | QKC98726 | CopG family transcriptional regulator | - |
| EB231_31870 | 6635623..6637653 | - | 2031 | QKC98727 | conjugal transfer protein TraG | - |
| EB231_31875 | 6637764..6638010 | + | 247 | Protein_6313 | XRE family transcriptional regulator | - |
| EB231_31880 | 6638309..6638740 | - | 432 | QKC98728 | hypothetical protein | - |
| EB231_31885 | 6638909..6639595 | + | 687 | QKC98729 | hypothetical protein | - |
Region 2: 6656071..6665494
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EB231_31945 | 6651676..6651803 | + | 128 | Protein_6327 | IS630 family transposase | - |
| EB231_31950 | 6651803..6652426 | + | 624 | Protein_6328 | ISKra4 family transposase | - |
| EB231_31955 | 6653408..6655948 | + | 2541 | QKC98737 | two-component system VirA-like sensor kinase | - |
| EB231_31960 | 6656071..6657111 | - | 1041 | QKC98738 | P-type DNA transfer ATPase VirB11 | virB11 |
| EB231_31965 | 6657095..6658234 | - | 1140 | QKC98739 | type IV secretion system protein VirB10 | virB10 |
| EB231_31970 | 6658231..6659112 | - | 882 | QKC98740 | type IV secretion system protein VirB9 | virB9 |
| EB231_31975 | 6659109..6659822 | - | 714 | QKC98741 | type IV secretion system protein VirB8 | virB8 |
| EB231_31980 | 6659819..6659977 | - | 159 | QKC98742 | type IV secretion system lipoprotein VirB7 | - |
| EB231_31985 | 6660004..6660891 | - | 888 | QKC98743 | type IV secretion system protein | virB6 |
| EB231_31990 | 6660949..6661623 | - | 675 | QKC98744 | type IV secretion system protein VirB5 | - |
| EB231_31995 | 6661637..6664006 | - | 2370 | QKC98745 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| EB231_32000 | 6664006..6664332 | - | 327 | QKC98746 | type IV secretion system protein VirB3 | virB3 |
| EB231_32005 | 6664332..6664697 | - | 366 | QKC98747 | pilin major subunit VirB2 | virB2 |
| EB231_32010 | 6664712..6665494 | - | 783 | QKC98748 | type IV secretion system lytic transglycosylase VirB1 | virB1 |
| EB231_32015 | 6666790..6667515 | + | 726 | QKC98749 | response regulator | - |
| EB231_32020 | 6667491..6668108 | - | 618 | QKC98750 | hypothetical protein | - |
Host bacterium
| ID | 394 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEMsp.SymNZP2298 | GenBank | CP033365 |
| Element size | 7336816 bp | Coordinate of oriT [Strand] | 146676..146735 [+] |
| Host bacterium | Mesorhizobium sp. NZP2298 | Coordinate of element | 6602423..7069563 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixS, fixI, fixH, fixG, fixP, fixQ, fixO, fixN, nodM, nodD, nolL, nodB, eB231_32485, nodJ, nodI, nodC, nodA, nodS, nifU, mlr6097, nifX, nifN, nifE, nifK, nifD, nifH, nifQ, mLTONO_5203, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, nifT, nolK, noeL, nodZ |
| Anti-CRISPR | AcrIIA9, AcrIIC1 |