Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200442 |
| Name | oriT_ICEMsp.SymNZP2234 |
| Organism | Mesorhizobium sp. NZP2234 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP033364 (118212..118272 [+], 61 nt) |
| oriT length | 61 nt |
| IRs (inverted repeats) | 27..33, 37..43 (CCGCCTC..GAGGCGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 61 nt
>oriT_ICEMsp.SymNZP2234
AAAACGATGTGCAGACAAAGGCTTCGCCGCCTCATGGAGGCGGTGCCTTTATCTTGCCTTA
AAAACGATGTGCAGACAAAGGCTTCGCCGCCTCATGGAGGCGGTGCCTTTATCTTGCCTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file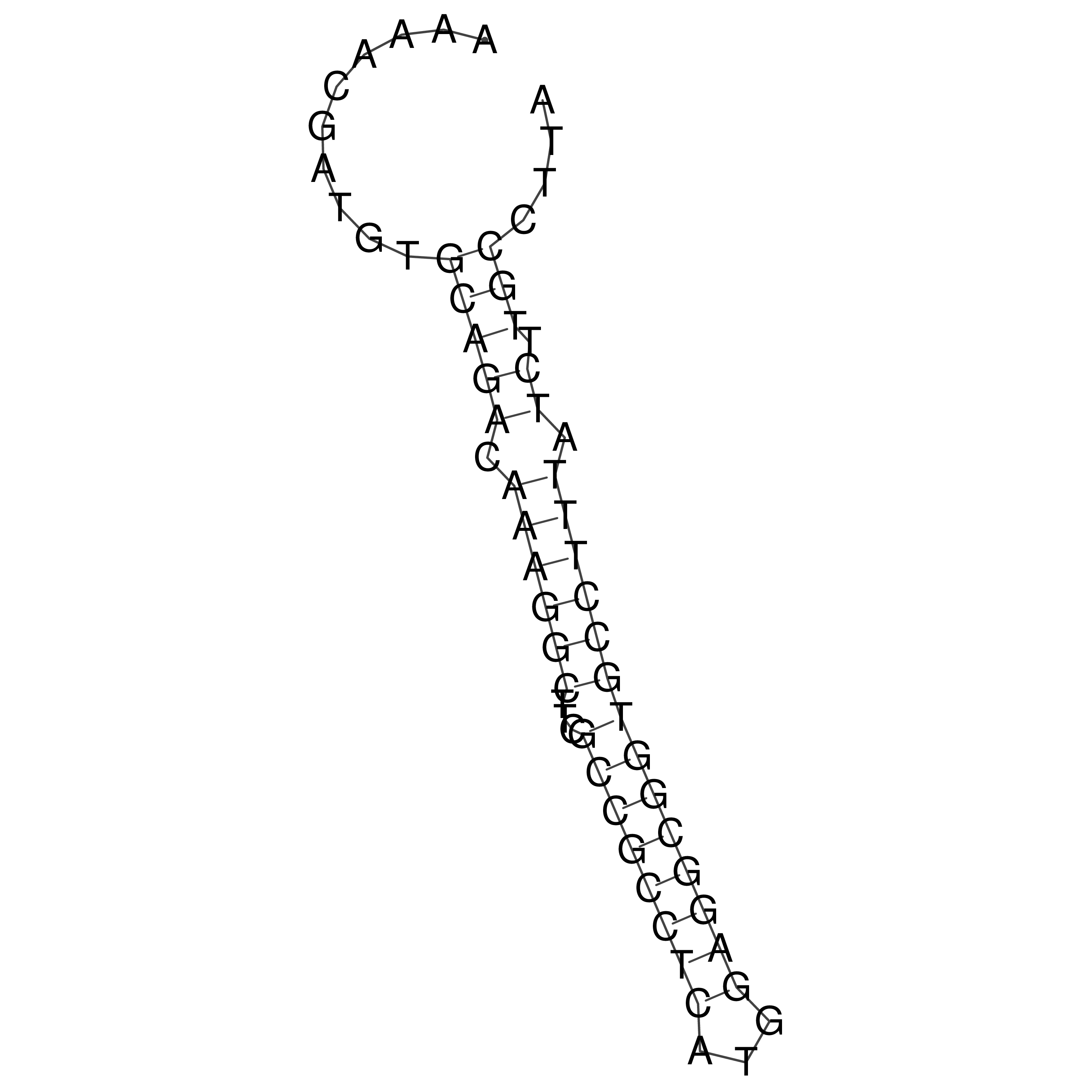
T4CP
| ID | 17035 | GenBank | QKC92218 |
| Name | t4cp2_EB230_30340_ICEMsp.SymNZP2234 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 90304.90 Da Isoelectric Point: 6.1180
>QKC92218.1 conjugal transfer protein TrbE [Mesorhizobium sp. NZP2234]
MLNLSEYRGKADRLADHLPWAALVAPGIVLNKDGSVQRTLRFRGPDLESSTEAELIGICARANDALRRLG
SGWTLFFEAERIEALGYPSSRFPDAASWLVDEERRAAFEGKVAHFESRYHLTLLSMPPPDAQARAESALV
DKHHSEGERDWRQEMARFRDETDRVLDLLSGFMPEVRPLDDAETLTYLHDTISTRRHPVAVPETPMYLDG
ILVDVPLTGGLEPMLGEQHLRTLTILGFPNLTRPGILDALNHQDFAYRWMTRFIPLGKTEATKTLTRLRR
QWFAKRKSIVAILREVVSNEPVPLIDSDADNKALDADEALQALGGDHVGFGYLTATVTVWDEDRQAAAEK
LRAVERIINGLGFTTIRESVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAEF
TGREAPPLLIAETSGSTPFRLSSHVEDVGHMLIVGPTGAGKSVLLALIALQFRRYAGAQVYVFDKGNSAR
AATLAMGGEHHVLGSDGALAFQPLRNIGDQGRRSWAAEWIAGLIAHENVTLTPEVKEAIWSALNSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHSQSAVLPVLTYLF
HRLEERFDGRPTLLMLDEAWVYLDNPLFAARIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQARTAYERFGLNDRQIELIARATPKRHYYLQSRCGNRLFELGLGPIALALCGASGPA
AQSLIDRVLSKHGQDSFVFRFLGARGLDWAAELLQQFPQPTKEQSS
MLNLSEYRGKADRLADHLPWAALVAPGIVLNKDGSVQRTLRFRGPDLESSTEAELIGICARANDALRRLG
SGWTLFFEAERIEALGYPSSRFPDAASWLVDEERRAAFEGKVAHFESRYHLTLLSMPPPDAQARAESALV
DKHHSEGERDWRQEMARFRDETDRVLDLLSGFMPEVRPLDDAETLTYLHDTISTRRHPVAVPETPMYLDG
ILVDVPLTGGLEPMLGEQHLRTLTILGFPNLTRPGILDALNHQDFAYRWMTRFIPLGKTEATKTLTRLRR
QWFAKRKSIVAILREVVSNEPVPLIDSDADNKALDADEALQALGGDHVGFGYLTATVTVWDEDRQAAAEK
LRAVERIINGLGFTTIRESVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAEF
TGREAPPLLIAETSGSTPFRLSSHVEDVGHMLIVGPTGAGKSVLLALIALQFRRYAGAQVYVFDKGNSAR
AATLAMGGEHHVLGSDGALAFQPLRNIGDQGRRSWAAEWIAGLIAHENVTLTPEVKEAIWSALNSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHSQSAVLPVLTYLF
HRLEERFDGRPTLLMLDEAWVYLDNPLFAARIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQARTAYERFGLNDRQIELIARATPKRHYYLQSRCGNRLFELGLGPIALALCGASGPA
AQSLIDRVLSKHGQDSFVFRFLGARGLDWAAELLQQFPQPTKEQSS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17036 | GenBank | QKC92223 |
| Name | t4cp2_EB230_30365_ICEMsp.SymNZP2234 |
UniProt ID | _ |
| Length | 676 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 676 a.a. Molecular weight: 75113.52 Da Isoelectric Point: 6.6899
>QKC92223.1 conjugal transfer protein TraG [Mesorhizobium sp. NZP2234]
MTPTKLLIGQIAIVFAIVLLGVWAATQWCAHMLAFQEQLGAPWFVAAGWPIYEPWKLLEWWFQFDAYAPE
VFDKAGMLAGTSGFMGCAAAIAGSLWRARQRGLVTTYGSSRWAATREIEKAGLFQPAGVFLGKLKDRYLR
HDGPEHVMAFAPTRSGKGVGLVVPTLLSWSGSAVIHDIKGENWQLSSGWRSRFSHCLLFNPTDPRSARYN
PLLEVRKGSDEIRDVQNIADILVDPEGALERRNHWEKTSHSLLVGAILHVLYAEEEKTLARVATFLSDPQ
RSFAATLRRMMTTNHLGAANKPQVHPVVASAAREVLNKSENERSGVLSTAMSFLGLYRDPTVAAVTSDCD
WRIADLMDAEWPMSLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPKKSRKHQLLMMLDEFPALGR
LDFFETALAFMAGYGIRAYLIAQSLNQISKAYGDNNAILDNCHLRIAFSSNDERTAKRISDALGTATELR
SMRNYAGHRLAPWLSHVMVSRQETARPLLTPGEVMQLPPADELVLVSGLPPIRAKKLRYYEDHNFTERVL
PSPVLRDGPYADCPMSRPDDWTGQVRGVDRRLASDDESAGAAHADEGGVQQQRHPGLPEEHTAVTQEPDQ
DDLSRPADDDSEAMADKRVMDRISTVAGAYGINEGRGDDKDIVPGF
MTPTKLLIGQIAIVFAIVLLGVWAATQWCAHMLAFQEQLGAPWFVAAGWPIYEPWKLLEWWFQFDAYAPE
VFDKAGMLAGTSGFMGCAAAIAGSLWRARQRGLVTTYGSSRWAATREIEKAGLFQPAGVFLGKLKDRYLR
HDGPEHVMAFAPTRSGKGVGLVVPTLLSWSGSAVIHDIKGENWQLSSGWRSRFSHCLLFNPTDPRSARYN
PLLEVRKGSDEIRDVQNIADILVDPEGALERRNHWEKTSHSLLVGAILHVLYAEEEKTLARVATFLSDPQ
RSFAATLRRMMTTNHLGAANKPQVHPVVASAAREVLNKSENERSGVLSTAMSFLGLYRDPTVAAVTSDCD
WRIADLMDAEWPMSLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPKKSRKHQLLMMLDEFPALGR
LDFFETALAFMAGYGIRAYLIAQSLNQISKAYGDNNAILDNCHLRIAFSSNDERTAKRISDALGTATELR
SMRNYAGHRLAPWLSHVMVSRQETARPLLTPGEVMQLPPADELVLVSGLPPIRAKKLRYYEDHNFTERVL
PSPVLRDGPYADCPMSRPDDWTGQVRGVDRRLASDDESAGAAHADEGGVQQQRHPGLPEEHTAVTQEPDQ
DDLSRPADDDSEAMADKRVMDRISTVAGAYGINEGRGDDKDIVPGF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 17037 | GenBank | QKC92246 |
| Name | t4cp2_EB230_30530_ICEMsp.SymNZP2234 |
UniProt ID | _ |
| Length | 563 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 563 a.a. Molecular weight: 62265.55 Da Isoelectric Point: 8.7896
>QKC92246.1 type IV secretion system protein VirD4 [Mesorhizobium sp. NZP2234]
MSSTKTTSPNIAISIACSLALGFCAASLYATFRHGGSGEALMAFDVRAFWFETPFYLGFATPVFYQGAAI
VLSTSAVVLLIQQFVLRRKLEHHGTARWARVDEMRRTGYLRRYRGVTGPVFGKTSGPFWPGYYLTNGEQP
HSLIVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKAAGHEVFKFAPLDSERRTNCYNPL
LDLIALPPQLQFTEARRLAANLITAKGEGAEGFVSGARDIFVAGILACIERGTPTIGAVYDLFAQPGEKF
KLFAQLAMETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPAVKAATSRSDFSVYDLRRRRTC
IYLCVGPNDLEVIAPLIRLFFQQTVSILQRSLPRQDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMLIIQSLSALTGAYGEAGKQNFLSNTGLQVFMATADDETPTYISKAIGEYTFEAKSISYSQSRAFDRNI
QNSFQGAPLLRPEQVRLLNDEYEIVLIKGLPPLQLRKVRYFSDRILKRIFESQTGALPEPAPLMEAHPDD
TDG
MSSTKTTSPNIAISIACSLALGFCAASLYATFRHGGSGEALMAFDVRAFWFETPFYLGFATPVFYQGAAI
VLSTSAVVLLIQQFVLRRKLEHHGTARWARVDEMRRTGYLRRYRGVTGPVFGKTSGPFWPGYYLTNGEQP
HSLIVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKAAGHEVFKFAPLDSERRTNCYNPL
LDLIALPPQLQFTEARRLAANLITAKGEGAEGFVSGARDIFVAGILACIERGTPTIGAVYDLFAQPGEKF
KLFAQLAMETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPAVKAATSRSDFSVYDLRRRRTC
IYLCVGPNDLEVIAPLIRLFFQQTVSILQRSLPRQDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMLIIQSLSALTGAYGEAGKQNFLSNTGLQVFMATADDETPTYISKAIGEYTFEAKSISYSQSRAFDRNI
QNSFQGAPLLRPEQVRLLNDEYEIVLIKGLPPLQLRKVRYFSDRILKRIFESQTGALPEPAPLMEAHPDD
TDG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 6230642..6239698
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EB230_30295 | 6227024..6227563 | + | 540 | QKC92210 | hemerythrin domain-containing protein | - |
| EB230_30300 | 6227567..6228322 | - | 756 | QKC92211 | Crp/Fnr family transcriptional regulator | - |
| EB230_30305 | 6228479..6229828 | + | 1350 | QKC92212 | oxygen-independent coproporphyrinogen III oxidase | - |
| EB230_30310 | 6230357..6230629 | - | 273 | QKC92213 | DUF2274 domain-containing protein | - |
| EB230_30315 | 6230642..6231856 | - | 1215 | QKC93249 | TrbI/VirB10 family protein | virB10 |
| EB230_30320 | 6231868..6232896 | - | 1029 | QKC92214 | P-type conjugative transfer protein TrbG | virB9 |
| EB230_30325 | 6232893..6233618 | - | 726 | QKC92215 | conjugal transfer protein TrbF | virB8 |
| EB230_30330 | 6233619..6234941 | - | 1323 | QKC92216 | P-type conjugative transfer protein TrbL | virB6 |
| EB230_30335 | 6234972..6235697 | - | 726 | QKC92217 | P-type conjugative transfer protein TrbJ | virB5 |
| EB230_30340 | 6235694..6238144 | - | 2451 | QKC92218 | conjugal transfer protein TrbE | virb4 |
| EB230_30345 | 6238138..6238410 | - | 273 | QKC92219 | conjugal transfer protein | virB3 |
| EB230_30350 | 6238410..6238718 | - | 309 | QKC92220 | conjugal transfer protein TrbC | virB2 |
| EB230_30355 | 6238730..6239698 | - | 969 | QKC92221 | P-type conjugative transfer ATPase TrbB | virB11 |
| EB230_30360 | 6239695..6240108 | - | 414 | QKC92222 | CopG family transcriptional regulator | - |
| EB230_30365 | 6240227..6242257 | - | 2031 | QKC92223 | conjugal transfer protein TraG | - |
| EB230_30370 | 6242387..6242650 | + | 264 | Protein_6013 | XRE family transcriptional regulator | - |
| EB230_30375 | 6242949..6243338 | - | 390 | QKC92224 | hypothetical protein | - |
Region 2: 6259951..6269374
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EB230_30430 | 6255556..6255683 | + | 128 | Protein_6025 | IS630 family transposase | - |
| EB230_30435 | 6255683..6256309 | + | 627 | Protein_6026 | ISKra4 family transposase | - |
| EB230_30440 | 6257288..6259831 | + | 2544 | QKC92232 | two-component system VirA-like sensor kinase | - |
| EB230_30445 | 6259951..6260991 | - | 1041 | QKC92233 | P-type DNA transfer ATPase VirB11 | virB11 |
| EB230_30450 | 6260975..6262114 | - | 1140 | QKC92234 | type IV secretion system protein VirB10 | virB10 |
| EB230_30455 | 6262111..6262992 | - | 882 | QKC92235 | type IV secretion system protein VirB9 | virB9 |
| EB230_30460 | 6262989..6263702 | - | 714 | QKC92236 | type IV secretion system protein VirB8 | virB8 |
| EB230_30465 | 6263699..6263857 | - | 159 | QKC92237 | type IV secretion system lipoprotein VirB7 | - |
| EB230_30470 | 6263884..6264771 | - | 888 | QKC92238 | type IV secretion system protein | virB6 |
| EB230_30475 | 6264829..6265503 | - | 675 | QKC92239 | type IV secretion system protein VirB5 | - |
| EB230_30480 | 6265517..6267886 | - | 2370 | QKC92240 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| EB230_30485 | 6267886..6268212 | - | 327 | QKC92241 | type IV secretion system protein VirB3 | virB3 |
| EB230_30490 | 6268212..6268577 | - | 366 | QKC92242 | pilin major subunit VirB2 | virB2 |
| EB230_30495 | 6268592..6269374 | - | 783 | QKC92243 | type IV secretion system lytic transglycosylase VirB1 | virB1 |
| EB230_30500 | 6270676..6271401 | + | 726 | QKC92244 | response regulator | - |
| EB230_30505 | 6271482..6271944 | - | 463 | Protein_6040 | IS66 family transposase | - |
Host bacterium
| ID | 393 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEMsp.SymNZP2234 | GenBank | CP033364 |
| Element size | 6749717 bp | Coordinate of oriT [Strand] | 118212..118272 [+] |
| Host bacterium | Mesorhizobium sp. NZP2234 | Coordinate of element | 6206258..6628681 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd, htpB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixS, fixI, fixH, fixG, fixP, fixQ, fixO, fixN, nodM, nodD, nolL, nodB, eB230_30840, nodJ, nodI, nodC, nodA, nodS, mlr6097, nifU, nifX, nifN, nifE, nifK, nifD, nifH, nifQ, mLTONO_5203, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, nifT, nolK, noeL, nodZ, nodF |
| Anti-CRISPR | AcrIIC1 |