Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200420 |
| Name | oriT_ICEMcSymWSM1497-α |
| Organism | Mesorhizobium sp. WSM1497 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP021070 (40888..40947 [+], 60 nt) |
| oriT length | 60 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 60 nt
>oriT_ICEMcSymWSM1497-α
AAAATGATGTGCAGACAGACACTTCGCCGGCCCTGGGAGGCGGTGCCTTTATCTTGCCTT
AAAATGATGTGCAGACAGACACTTCGCCGGCCCTGGGAGGCGGTGCCTTTATCTTGCCTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file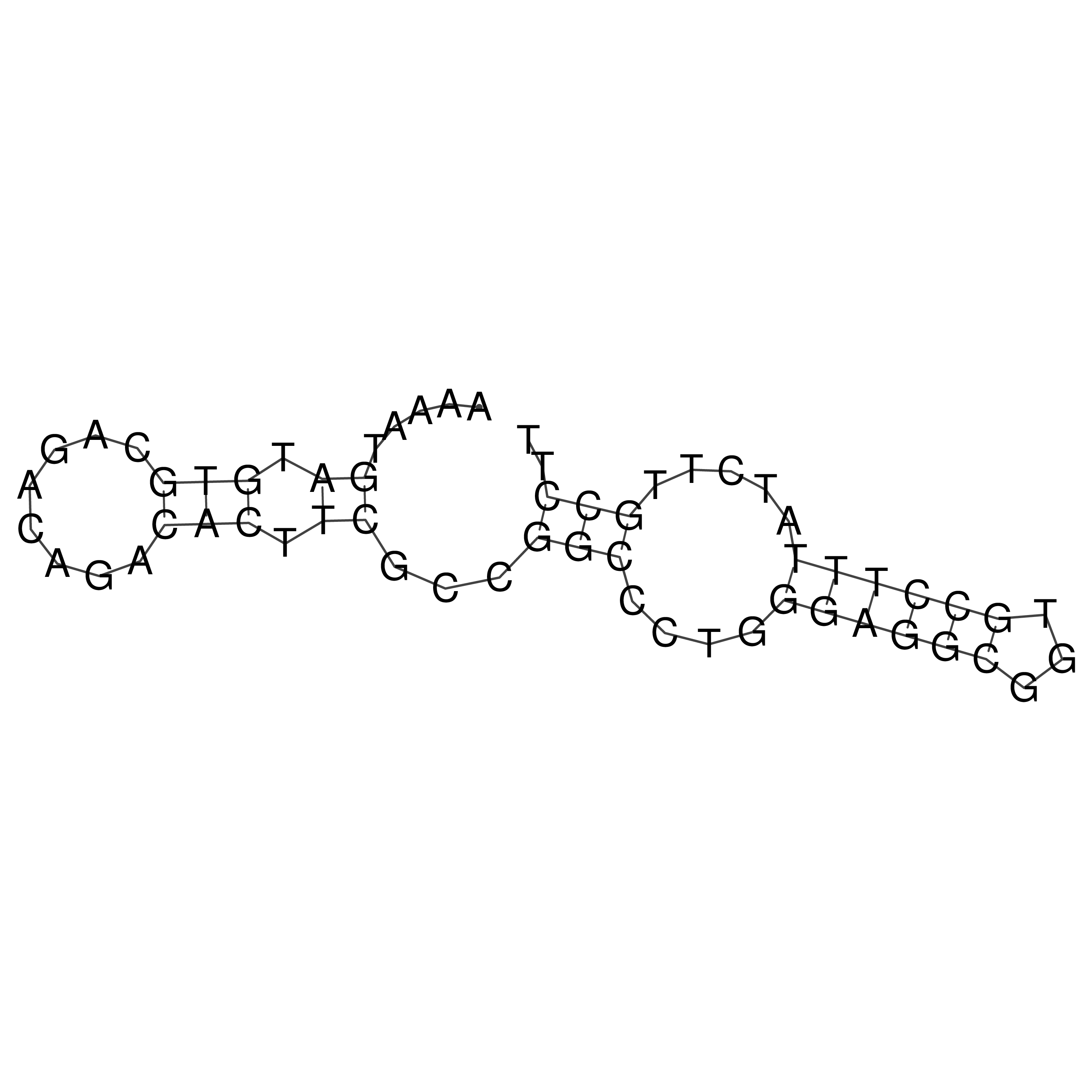
T4CP
| ID | 16987 | GenBank | ARP67114 |
| Name | t4cp2_A9K65_030005_ICEMcSymWSM1497-α |
UniProt ID | _ |
| Length | 813 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 813 a.a. Molecular weight: 89718.33 Da Isoelectric Point: 6.6912
>ARP67114.1 conjugal transfer protein TrbE [Mesorhizobium sp. WSM1497]
MLNLSEYRGKADRLADHLPWAALVAPGIVLNKDGSFQRSFGFRGPDLESATEAELIGICARANNALRRLG
SGWALFFDAERIEALGYPSSRFPDAASWLVDEERRAAFQGKVAHYESRYHLTLTFIPPPDSQARAQSALV
DSHHSGGEREWRQELTRFRDETGRVLDLLSGFMPEIRALDDAGTLTYLHGTISTRRHPVAAPKTPMYLDG
ILVDAPLTGGLEPMLGNHHLRTLTILGFPNVTRPGILDALNNQDFPYRWMTRFIPLDKTEATRTLTRLRR
QWFAKRKSIVAILREVVTSEPVPLVDTDAENKALDADAAIQALGGDHVGFGYLTTTVTVWDEDHQAAAEK
LRAVERVVNGLGFATIREGVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAKV
TQAEAPPLLFASTSGSTPFRLSTHVDDVGHMLIVGPTGAGKSVLVALFALQFRRYAGAQVYVFDKGNSAR
AAILAMGGEHHALRAHGALAFQPLRKIDDPAIRSWAAEWIAGLFAHENVTVTPEAKEAIWSALTSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHSQGAVLPVLTYLF
HRLEARFDGRPTLLMLDEAWLYLDNPLFAGRIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQAREAYERFGLNERQIELVARATPKRQYYLQSRRGNRLFQLGLGPIALALCGASDPA
SQTLIDKILSERGQDGFAFYFLNARGLDWAAELLRQFPQPDME
MLNLSEYRGKADRLADHLPWAALVAPGIVLNKDGSFQRSFGFRGPDLESATEAELIGICARANNALRRLG
SGWALFFDAERIEALGYPSSRFPDAASWLVDEERRAAFQGKVAHYESRYHLTLTFIPPPDSQARAQSALV
DSHHSGGEREWRQELTRFRDETGRVLDLLSGFMPEIRALDDAGTLTYLHGTISTRRHPVAAPKTPMYLDG
ILVDAPLTGGLEPMLGNHHLRTLTILGFPNVTRPGILDALNNQDFPYRWMTRFIPLDKTEATRTLTRLRR
QWFAKRKSIVAILREVVTSEPVPLVDTDAENKALDADAAIQALGGDHVGFGYLTTTVTVWDEDHQAAAEK
LRAVERVVNGLGFATIREGVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAKV
TQAEAPPLLFASTSGSTPFRLSTHVDDVGHMLIVGPTGAGKSVLVALFALQFRRYAGAQVYVFDKGNSAR
AAILAMGGEHHALRAHGALAFQPLRKIDDPAIRSWAAEWIAGLFAHENVTVTPEAKEAIWSALTSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHSQGAVLPVLTYLF
HRLEARFDGRPTLLMLDEAWLYLDNPLFAGRIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQAREAYERFGLNERQIELVARATPKRQYYLQSRRGNRLFQLGLGPIALALCGASDPA
SQTLIDKILSERGQDGFAFYFLNARGLDWAAELLRQFPQPDME
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16988 | GenBank | ARP67119 |
| Name | t4cp2_A9K65_030030_ICEMcSymWSM1497-α |
UniProt ID | _ |
| Length | 676 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 676 a.a. Molecular weight: 75131.58 Da Isoelectric Point: 7.0969
>ARP67119.1 conjugal transfer protein TraG [Mesorhizobium sp. WSM1497]
MTPTKLLIGQIVVVFAIVMLGVWTATQWCAHMLGHQPPLGAPWFVAAGWPIYKPWKLFEWWYHFDAYAPE
VFDKAGALAGASGFVGCAAAIGGSLWRARQLGSVTTYGSSRWATTQEIVTAGLFRPAGVFLGKLKDRYLR
HDGPEHVMAFAPTRSGKGVGLVVPTLLSWTGSAVIHDIKGENWQLTSGWRSKFSYCLLFNPTDPRSARYN
PLLEVRKGPQEVRDVQNIADILVDPEGALDRRNHWEKTSHSLLVGAVLHVLYAEEEKTLARVATFLSDPQ
RSFSATLQQMMTTNHLGTPDKPEVHPVVASAARELLNKSENERSGVLSTAMSFLGLYRDPTVAAATSTCD
WRIVDLMDAERPISLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPSRSRKHRLLMMLDEFPALGR
LDFFETALAFMAGYGIRAYLIAQSLNQISKAYGENSAILDNCHVRIAFSSNDERTAKRISDALGTATELR
SMRNYAGHRLAPWLSHVMVSRQETARPLLTPGEVMQLPQTDELVLISGLPPLRVQKLRYYEDRNFTERVL
PSPVLRDSPYADRPASRTDDWTGRVRSVDGRLAAGDEIADAAIGDEGGVQQQRHPALPEHQIVLSQEAEQ
DDLFKSVDDDSEPVADKRVMDRTSTAARVYGLNEGRGDDKDIVPGF
MTPTKLLIGQIVVVFAIVMLGVWTATQWCAHMLGHQPPLGAPWFVAAGWPIYKPWKLFEWWYHFDAYAPE
VFDKAGALAGASGFVGCAAAIGGSLWRARQLGSVTTYGSSRWATTQEIVTAGLFRPAGVFLGKLKDRYLR
HDGPEHVMAFAPTRSGKGVGLVVPTLLSWTGSAVIHDIKGENWQLTSGWRSKFSYCLLFNPTDPRSARYN
PLLEVRKGPQEVRDVQNIADILVDPEGALDRRNHWEKTSHSLLVGAVLHVLYAEEEKTLARVATFLSDPQ
RSFSATLQQMMTTNHLGTPDKPEVHPVVASAARELLNKSENERSGVLSTAMSFLGLYRDPTVAAATSTCD
WRIVDLMDAERPISLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPSRSRKHRLLMMLDEFPALGR
LDFFETALAFMAGYGIRAYLIAQSLNQISKAYGENSAILDNCHVRIAFSSNDERTAKRISDALGTATELR
SMRNYAGHRLAPWLSHVMVSRQETARPLLTPGEVMQLPQTDELVLISGLPPLRVQKLRYYEDRNFTERVL
PSPVLRDSPYADRPASRTDDWTGRVRSVDGRLAAGDEIADAAIGDEGGVQQQRHPALPEHQIVLSQEAEQ
DDLFKSVDDDSEPVADKRVMDRTSTAARVYGLNEGRGDDKDIVPGF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16989 | GenBank | ARP67183 |
| Name | t4cp2_A9K65_030425_ICEMcSymWSM1497-α |
UniProt ID | _ |
| Length | 581 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 581 a.a. Molecular weight: 64321.88 Da Isoelectric Point: 7.9553
>ARP67183.1 type IV secretion system protein VirD4 [Mesorhizobium sp. WSM1497]
MPSTKTTTPNIAVSIACSLAVGFCAASLYATFLHGGGGEALMAFDVRAFWFETPFYLGFATPVFYRGAAI
VLSTSAVVLLIQQVVLRRKLEHHGTARWARVDEMRRTGYLRRYRGVKGPVFGKTSGPFWPGYYLTNGEQP
HSLIVAPTRAGKGVGVVIPTLLTFKGSVIALDVKGELFELTSRARKAAGDDVFKFAPLDSERRTNCYNPL
LDLIALPPQLQFTEARRLAANLITAKGEGAEGFVSGARDIFVAGILACIERGTPTIGAVYDLFAQPGEKF
KLFAQLAMETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPAVKAATSRSDFSVYDLRRRRTC
IYLCVGPNDLEVIAPLIRLFFQQIVSILQRSLPRPEEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMLIIQSLSALTGAYGEAGKQNFLSNTGLQVFMATADDETPNYISKAIGEYTFEAKSISYSQARRFDWNI
QNSHQGAPLLRPEQVRLLDDDYEIVLIKGLPPLQLRKVRYFSDRILKRIFEGQTGALPEPAPLTEAEEEL
STDFRLDEPPRGLAQEFNDTV
MPSTKTTTPNIAVSIACSLAVGFCAASLYATFLHGGGGEALMAFDVRAFWFETPFYLGFATPVFYRGAAI
VLSTSAVVLLIQQVVLRRKLEHHGTARWARVDEMRRTGYLRRYRGVKGPVFGKTSGPFWPGYYLTNGEQP
HSLIVAPTRAGKGVGVVIPTLLTFKGSVIALDVKGELFELTSRARKAAGDDVFKFAPLDSERRTNCYNPL
LDLIALPPQLQFTEARRLAANLITAKGEGAEGFVSGARDIFVAGILACIERGTPTIGAVYDLFAQPGEKF
KLFAQLAMETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPAVKAATSRSDFSVYDLRRRRTC
IYLCVGPNDLEVIAPLIRLFFQQIVSILQRSLPRPEEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMLIIQSLSALTGAYGEAGKQNFLSNTGLQVFMATADDETPNYISKAIGEYTFEAKSISYSQARRFDWNI
QNSHQGAPLLRPEQVRLLDDDYEIVLIKGLPPLQLRKVRYFSDRILKRIFEGQTGALPEPAPLTEAEEEL
STDFRLDEPPRGLAQEFNDTV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 6125437..6134498
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| A9K65_029960 | 6120814..6122442 | - | 1629 | ARP67105 | cytochrome-c oxidase, cbb3-type subunit I | - |
| A9K65_029965 | 6122779..6123528 | - | 750 | ARP67106 | transcriptional regulator | - |
| A9K65_029970 | 6123707..6125029 | + | 1323 | ARP67107 | oxygen-independent coproporphyrinogen III oxidase | - |
| A9K65_029975 | 6125162..6125434 | - | 273 | ARP67108 | transposase | - |
| A9K65_029980 | 6125437..6126657 | - | 1221 | ARP67109 | conjugal transfer protein TrbI | virB10 |
| A9K65_029985 | 6126671..6127699 | - | 1029 | ARP67110 | P-type conjugative transfer protein TrbG | virB9 |
| A9K65_029990 | 6127689..6128420 | - | 732 | ARP67111 | conjugal transfer protein TrbF | virB8 |
| A9K65_029995 | 6128421..6129743 | - | 1323 | ARP67112 | P-type conjugative transfer protein TrbL | virB6 |
| A9K65_030000 | 6129772..6130497 | - | 726 | ARP67113 | P-type conjugative transfer protein TrbJ | virB5 |
| A9K65_030005 | 6130503..6132944 | - | 2442 | ARP67114 | conjugal transfer protein TrbE | virb4 |
| A9K65_030010 | 6132938..6133210 | - | 273 | ARP67115 | conjugal transfer protein | virB3 |
| A9K65_030015 | 6133210..6133518 | - | 309 | ARP67116 | conjugal transfer protein TrbC | virB2 |
| A9K65_030020 | 6133530..6134498 | - | 969 | ARP67117 | P-type conjugative transfer ATPase TrbB | virB11 |
| A9K65_030025 | 6134495..6134908 | - | 414 | ARP67118 | CopG family transcriptional regulator | - |
| A9K65_030030 | 6135025..6137055 | - | 2031 | ARP67119 | conjugal transfer protein TraG | - |
| A9K65_030035 | 6137256..6139079 | - | 1824 | ARP67120 | glutamine-fructose-6-phosphate transaminase (isomerizing) | - |
Region 2: 6200123..6209546
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| A9K65_030335 | 6196649..6197401 | + | 753 | ARP67169 | carbonate dehydratase | - |
| A9K65_030340 | 6197569..6200004 | + | 2436 | ARP67170 | sensor histidine kinase | - |
| A9K65_030345 | 6200123..6201163 | - | 1041 | ARP67171 | P-type DNA transfer ATPase VirB11 | virB11 |
| A9K65_030350 | 6201147..6202289 | - | 1143 | ARP67172 | type IV secretion system protein VirB10 | virB10 |
| A9K65_030355 | 6202286..6203167 | - | 882 | ARP67173 | type IV secretion system protein VirB9 | virB9 |
| A9K65_030360 | 6203164..6203877 | - | 714 | ARP67174 | type IV secretion system protein VirB8 | virB8 |
| A9K65_030365 | 6203874..6204032 | - | 159 | ARP67175 | type IV secretion system protein VirB7 | - |
| A9K65_030370 | 6204055..6204942 | - | 888 | ARP67176 | type IV secretion system protein VirB6 | virB6 |
| A9K65_030375 | 6205001..6205684 | - | 684 | ARP67177 | type IV secretion system protein VirB5 | - |
| A9K65_030380 | 6205698..6208067 | - | 2370 | ARP67178 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| A9K65_030385 | 6208067..6208393 | - | 327 | ARP67179 | type IV secretion system protein VirB3 | virB3 |
| A9K65_030390 | 6208393..6208758 | - | 366 | ARP67180 | type IV secretion system protein VirB2 | virB2 |
| A9K65_030395 | 6208773..6209546 | - | 774 | ARP67181 | lytic transglycosylase | virB1 |
| A9K65_030400 | 6209985..6210544 | + | 560 | Protein_6018 | hypothetical protein | - |
| A9K65_030405 | 6210769..6211494 | + | 726 | ARP68089 | DNA-binding response regulator | - |
| A9K65_030410 | 6211291..6211848 | - | 558 | ARP67182 | hypothetical protein | - |
| A9K65_030415 | 6211897..6213283 | - | 1387 | Protein_6021 | IS66 family transposase | - |
Host bacterium
| ID | 375 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEMcSymWSM1497-α | GenBank | CP021070 |
| Element size | 6666492 bp | Coordinate of oriT [Strand] | 40888..40947 [+] |
| Host bacterium | Mesorhizobium sp. WSM1497 | Coordinate of element | 6100974..6544486 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixS, fixI, fixH, fixG, fixP, fixQ, fixO, fixN, nodM, mlr6097, nifX, nifN, nifE, nifK, nifD, nifH, nifQ, nifS, nifW, fixA, fixB, fixC, fixX, nifB, nifZ, nifT, nifA, nodD, nodA, nodF, nodE, nodB, nodC, nodI, nodJ, nodH |
| Anti-CRISPR | - |