Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200412 |
| Name | oriT_Tn6815 |
| Organism | Streptococcus mitis |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | LR828204 (2501..2633 [+], 133 nt) |
| oriT length | 133 nt |
| IRs (inverted repeats) | 65..70, 83..88 (AAATCC..GGATTT) 6..12, 23..29 (ACCCCCC..GGGGGGT) |
| Location of nic site | 75..76 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 133 nt
>oriT_Tn6815
ACTTAACCCCCCGTTTCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
ACTTAACCCCCCGTTTCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file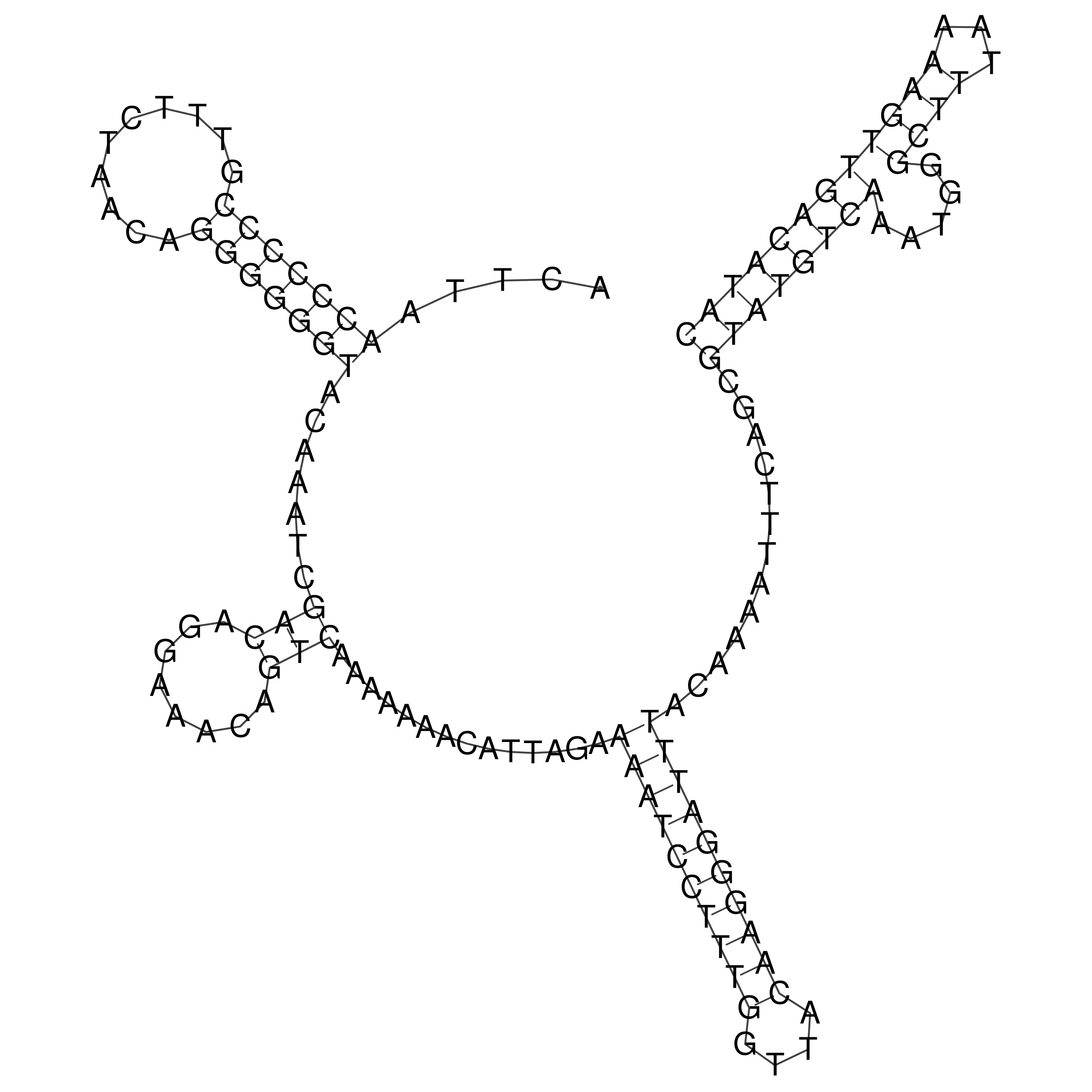
Relaxase
| ID | 14440 | GenBank | CAD0296864 |
| Name | Rep_trans_-_Tn6815 |
UniProt ID | _ |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
>CAD0296864.1 hypothetical protein [Streptococcus mitis]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7888 | GenBank | CAD0296881 |
| Name | CAD0296881_Tn6815 |
UniProt ID | _ |
| Length | 405 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 405 a.a. Molecular weight: 47021.92 Da Isoelectric Point: 9.7978
>CAD0296881.1 Transposase from transposon Tn916 [Streptococcus mitis]
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16973 | GenBank | CAD0296863 |
| Name | ftsK_2_-_Tn6815 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>CAD0296863.1 DNA translocase FtsK [Streptococcus mitis]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 336..11746
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_0 | 336..650 | + | 315 | CAD0296861 | hypothetical protein | orf23 |
| Locus_1 | 669..1052 | + | 384 | CAD0296862 | hypothetical protein | orf23 |
| Locus_2 | 1081..2466 | + | 1386 | CAD0296863 | DNA translocase FtsK | virb4 |
| Locus_3 | 2644..3849 | + | 1206 | CAD0296864 | hypothetical protein | - |
| Locus_4 | 3892..4113 | + | 222 | CAD0296865 | hypothetical protein | orf19 |
| Locus_5 | 4230..4727 | + | 498 | CAD0296866 | hypothetical protein | - |
| Locus_6 | 4702..5208 | + | 507 | CAD0296867 | hypothetical protein | orf17a |
| Locus_7 | 5192..7639 | + | 2448 | CAD0296868 | hypothetical protein | virb4 |
| Locus_8 | 7708..9819 | + | 2112 | CAD0296869 | hypothetical protein | orf15 |
| Locus_9 | 9816..10817 | + | 1002 | CAD0296870 | putative endopeptidase p60 | orf14 |
| Locus_10 | 10832..11746 | + | 915 | CAD0296871 | hypothetical protein | orf13 |
| Locus_11 | 12123..14042 | + | 1920 | CAD0296872 | Tetracycline resistance protein TetM from transposon TnFO1 | - |
| Locus_12 | 15149..15886 | + | 738 | CAD0296873 | rRNA adenine N-6-methyltransferase | - |
| Locus_13 | 15891..16022 | + | 132 | CAD0296874 | hypothetical protein | - |
Host bacterium
| ID | 367 | Element type | ICE (Integrative and conjugative element) |
| Element name | Tn6815 | GenBank | LR828204 |
| Element size | 23299 bp | Coordinate of oriT [Strand] | 2501..2633 [+] |
| Host bacterium | Streptococcus mitis | Coordinate of element | 1..23299 |
Cargo genes
| Drug resistance gene | tet(M), erm(B) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |