Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200406 |
| Name | oriT_ICEVpaChn0574 |
| Organism | Vibrio parahaemolyticus strain Vb0574 ICEVb0574 mobile element |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | MN199028 (3361..3658 [+], 298 nt) |
| oriT length | 298 nt |
| IRs (inverted repeats) | 44..49, 51..56 (CAAACG..CGTTTG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 298 nt
>oriT_ICEVpaChn0574
GCTCTGTTTGGCGGCGGATGACCTAGTCAAAAAATCGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTCGCCAAACGGTTTGTATCTTCATGACGATACGTCGTTTTAGGCGTTTTTAAGTGAAATCGGGCTGTATCCCTTGTCAGGTATGGGATTGCGCGAGTTGATTTATATCGAGACGCCAAACAGCGATTGTTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
GCTCTGTTTGGCGGCGGATGACCTAGTCAAAAAATCGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTCGCCAAACGGTTTGTATCTTCATGACGATACGTCGTTTTAGGCGTTTTTAAGTGAAATCGGGCTGTATCCCTTGTCAGGTATGGGATTGCGCGAGTTGATTTATATCGAGACGCCAAACAGCGATTGTTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file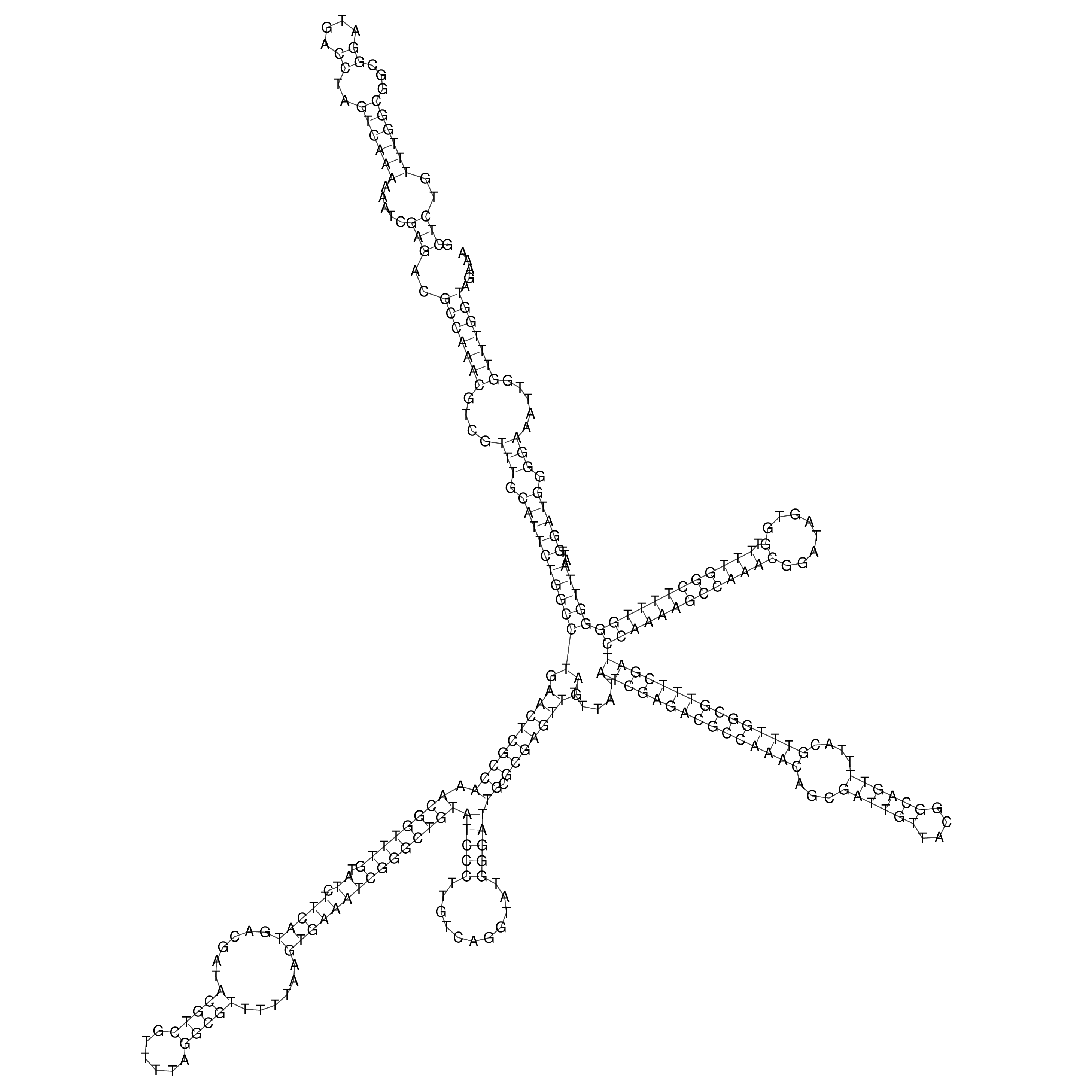
Relaxase
| ID | 14434 | GenBank | QIB99820 |
| Name | TraI_2_-_ICEVpaChn0574 |
UniProt ID | _ |
| Length | 716 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 716 a.a. Molecular weight: 79734.70 Da Isoelectric Point: 5.1503
>QIB99820.1 hypothetical protein [Vibrio parahaemolyticus]
MFKNLFFQAKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDENGLYQWNPFLETLSQWTTNSSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWVRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLPMLHISEADLLFRSNVPSSVRLFSKSEWEATQQTQAEPQSRSSEHSDL
PEASSSIEHGNSAESSSTNSSEQDDELRLASDVNNPQANENALGDGCEKPNNSYDGAISNNVNQHDAEAL
NLPESLAWLPEASSALVMVDEQILIRYPDAVRHWCAPRKLLAELSRLDWLELDPANPTRKARTVTTNDGV
QEQGLLLKVSISKGLSALIDLPKQDAESAAAIQNEEASQRPSRTKITNAQAKEPATRAERNQKPIALNAN
SITDPKHAQRQKMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAER
MFKNLFFQAKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDENGLYQWNPFLETLSQWTTNSSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWVRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLPMLHISEADLLFRSNVPSSVRLFSKSEWEATQQTQAEPQSRSSEHSDL
PEASSSIEHGNSAESSSTNSSEQDDELRLASDVNNPQANENALGDGCEKPNNSYDGAISNNVNQHDAEAL
NLPESLAWLPEASSALVMVDEQILIRYPDAVRHWCAPRKLLAELSRLDWLELDPANPTRKARTVTTNDGV
QEQGLLLKVSISKGLSALIDLPKQDAESAAAIQNEEASQRPSRTKITNAQAKEPATRAERNQKPIALNAN
SITDPKHAQRQKMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16966 | GenBank | QIB99821 |
| Name | t4cp2_-_ICEVpaChn0574 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67068.91 Da Isoelectric Point: 6.6444
>QIB99821.1 hypothetical protein [Vibrio parahaemolyticus]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQKG
LKGKPLAFMALTHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKNAMAMLIFYYERIQPIAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQKG
LKGKPLAFMALTHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKNAMAMLIFYYERIQPIAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16966 | GenBank | QIB99821 |
| Name | t4cp2_-_ICEVpaChn0574 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67068.91 Da Isoelectric Point: 6.6444
>QIB99821.1 hypothetical protein [Vibrio parahaemolyticus]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQKG
LKGKPLAFMALTHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKNAMAMLIFYYERIQPIAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQKG
LKGKPLAFMALTHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKNAMAMLIFYYERIQPIAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 23708..50785
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_14 | 20172..21353 | + | 1182 | QIB99819 | Endonuclease NucS | - |
| Locus_15 | 21510..23660 | + | 2151 | QIB99820 | hypothetical protein | - |
| Locus_16 | 23708..25528 | + | 1821 | QIB99821 | hypothetical protein | virb4 |
| Locus_17 | 25538..26098 | + | 561 | QIB99822 | hypothetical protein | - |
| Locus_18 | 26085..26720 | + | 636 | QIB99823 | hypothetical protein | tfc7 |
| Locus_19 | 27141..28403 | + | 1263 | QIB99824 | hypothetical protein | - |
| Locus_20 | 28653..29528 | + | 876 | QIB99825 | Beta-lactamase CTX-M-14 | - |
| Locus_21 | 29869..30399 | - | 531 | QIB99826 | hypothetical protein | - |
| Locus_22 | 30688..30969 | + | 282 | QIB99827 | hypothetical protein | traL |
| Locus_23 | 30966..31592 | + | 627 | QIB99828 | hypothetical protein | traE |
| Locus_24 | 31573..32472 | + | 900 | QIB99829 | hypothetical protein | traK |
| Locus_25 | 32475..33764 | + | 1290 | QIB99830 | hypothetical protein | traB |
| Locus_26 | 33839..34411 | + | 573 | QIB99831 | hypothetical protein | traV |
| Locus_27 | 34408..34794 | + | 387 | QIB99832 | hypothetical protein | - |
| Locus_28 | 34973..35806 | + | 834 | QIB99833 | hypothetical protein | - |
| Locus_29 | 35799..36527 | + | 729 | QIB99834 | hypothetical protein | - |
| Locus_30 | 36648..37259 | + | 612 | QIB99835 | hypothetical protein | - |
| Locus_31 | 37565..37783 | + | 219 | QIB99836 | hypothetical protein | - |
| Locus_32 | 37921..39138 | - | 1218 | QIB99837 | Tetracycline resistance protein | - |
| Locus_33 | 39219..39851 | + | 633 | QIB99838 | Tetracycline repressor protein | - |
| Locus_34 | 40336..40884 | + | 549 | QIB99839 | hypothetical protein | - |
| Locus_35 | 41016..41708 | + | 693 | QIB99840 | Thiol:disulfide interchange protein | trbB |
| Locus_36 | 41708..44107 | + | 2400 | QIB99841 | hypothetical protein | virb4 |
| Locus_37 | 44100..44447 | + | 348 | QIB99842 | hypothetical protein | - |
| Locus_38 | 44431..44943 | + | 513 | QIB99843 | hypothetical protein | - |
| Locus_39 | 44954..46078 | + | 1125 | QIB99844 | hypothetical protein | traW |
| Locus_40 | 46062..47090 | + | 1029 | QIB99845 | hypothetical protein | traU |
| Locus_41 | 47093..50785 | + | 3693 | QIB99846 | hypothetical protein | traN |
| Locus_42 | 51016..51348 | + | 333 | QIB99847 | hypothetical protein | - |
| Locus_43 | 51379..52041 | + | 663 | QIB99848 | hypothetical protein | - |
| Locus_44 | 52132..52734 | - | 603 | QIB99849 | hypothetical protein | - |
| Locus_45 | 53104..53430 | + | 327 | QIB99850 | hypothetical protein | - |
| Locus_46 | 53446..53865 | + | 420 | QIB99851 | Single-stranded DNA-binding protein | - |
| Locus_47 | 53945..54763 | + | 819 | QIB99852 | hypothetical protein | - |
| Locus_48 | 54845..54988 | + | 144 | QIB99853 | hypothetical protein | - |
Host bacterium
| ID | 361 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEVpaChn0574 | GenBank | MN199028 |
| Element size | 76762 bp | Coordinate of oriT [Strand] | 3361..3658 [+] |
| Host bacterium | Vibrio parahaemolyticus strain Vb0574 ICEVb0574 mobile element | Coordinate of element | 1..76762 |
Cargo genes
| Drug resistance gene | blaCTX-M-14, tet(E) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |