Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200405 |
| Name | oriT_ICEVchThd1 |
| Organism | Vibrio cholerae strain TSY216 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP007653 (5166..5464 [+], 299 nt) |
| oriT length | 299 nt |
| IRs (inverted repeats) | 234..242, 254..262 (AAAGCCAAA..TTTGGCTTT) 45..50, 52..57 (CAAACG..CGTTTG) 7..12, 26..31 (TTTGGC..GCCAAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 299 nt
>oriT_ICEVchThd1
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file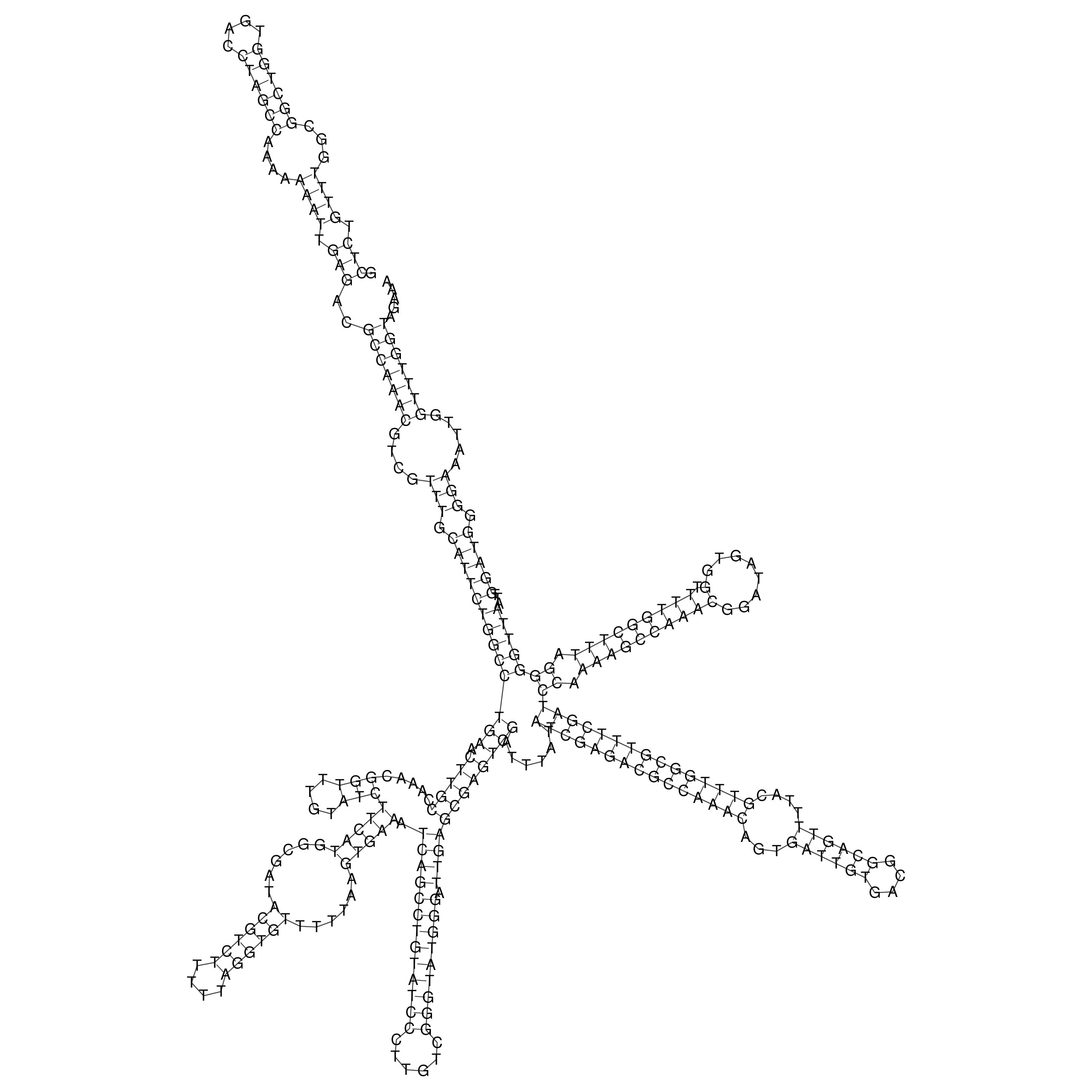
Relaxase
| ID | 14433 | GenBank | AKO74217 |
| Name | TraI_2_EN12_02990_ICEVchThd1 |
UniProt ID | _ |
| Length | 716 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 716 a.a. Molecular weight: 79789.91 Da Isoelectric Point: 5.2251
>AKO74217.1 phosphohydrolase [Vibrio cholerae]
MFKNLFFQTKALPELSSQLDTEIPRYPPFLKGLPAASPEDLQSTQDELITKLRQVLGFNLRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNTIGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTNLYLVWKSAAKEIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDGKPIWLPMLHISEADLLFSSNVPSGVRLFSKSEWEATQQTQAEPQSRSSEQPGL
SEASSSIELNNSAESPSTKSSEQDDERRLASDVNHLQATENAPGDECEKPNNSYDGAISNNVNQHDAEAL
NLPESLAWLPEASSALVMVDEQILVRYPDAVRHWCAPRKLLAELSRLDWLEIDPANPTRKARTVTTKGGV
QEQGLLLKVSISKGLTALIDLPKQDAESAAAIQNEEASPRPSRTETTNARAKEPSKRAECKQKPIAPNAN
SSIDPKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAER
MFKNLFFQTKALPELSSQLDTEIPRYPPFLKGLPAASPEDLQSTQDELITKLRQVLGFNLRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNTIGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTNLYLVWKSAAKEIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDGKPIWLPMLHISEADLLFSSNVPSGVRLFSKSEWEATQQTQAEPQSRSSEQPGL
SEASSSIELNNSAESPSTKSSEQDDERRLASDVNHLQATENAPGDECEKPNNSYDGAISNNVNQHDAEAL
NLPESLAWLPEASSALVMVDEQILVRYPDAVRHWCAPRKLLAELSRLDWLEIDPANPTRKARTVTTKGGV
QEQGLLLKVSISKGLTALIDLPKQDAESAAAIQNEEASPRPSRTETTNARAKEPSKRAECKQKPIAPNAN
SSIDPKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16964 | GenBank | AKO74218 |
| Name | t4cp2_EN12_02995_ICEVchThd1 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67134.99 Da Isoelectric Point: 6.4440
>AKO74218.1 conjugal transfer protein [Vibrio cholerae]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQMG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPERSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQMG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPERSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16965 | GenBank | AKO74234 |
| Name | t4cp2_EN12_03085_ICEVchThd1 |
UniProt ID | _ |
| Length | 799 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 799 a.a. Molecular weight: 90098.71 Da Isoelectric Point: 6.2880
>AKO74234.1 conjugal transfer protein TraC [Vibrio cholerae]
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAAISAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTGEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRNSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAAISAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTGEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRNSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 627884..652021
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EN12_02975 | 623863..624717 | + | 855 | AKO74214 | restriction endonuclease | - |
| EN12_02980 | 624811..625092 | + | 282 | AKO74215 | hypothetical protein | - |
| EN12_02985 | 625092..625565 | + | 474 | AKO74216 | hypothetical protein | - |
| EN12_02990 | 625685..627835 | + | 2151 | AKO74217 | phosphohydrolase | - |
| EN12_02995 | 627884..629704 | + | 1821 | AKO74218 | conjugal transfer protein | virb4 |
| EN12_03000 | 629714..630274 | + | 561 | AKO74219 | hypothetical protein | - |
| EN12_03005 | 630261..630896 | + | 636 | AKO74220 | conjugal transfer protein | tfc7 |
| EN12_03010 | 631168..632724 | + | 1557 | AKO74221 | transposase | - |
| EN12_03020 | 632884..633132 | + | 249 | AKO74222 | hypothetical protein | - |
| EN12_03025 | 633092..634363 | + | 1272 | AKO74223 | transposase | - |
| EN12_03030 | 634439..635134 | + | 696 | AKO74224 | ATPase AAA | virB11 |
| EN12_03035 | 635213..635785 | - | 573 | AKO74225 | hypothetical protein | - |
| EN12_03040 | 636074..636355 | + | 282 | AKO74226 | conjugal transfer protein TraL | traL |
| EN12_03045 | 636352..636978 | + | 627 | AKO74227 | pilus assembly protein | traE |
| EN12_03050 | 637052..637858 | + | 807 | AKO74228 | hypothetical protein | traK |
| EN12_03055 | 637861..639150 | + | 1290 | AKO74229 | conjugal transfer protein TraB | traB |
| EN12_03060 | 639147..639797 | + | 651 | AKO74230 | conjugal transfer protein TraV | traV |
| EN12_03065 | 639794..640180 | + | 387 | AKO74231 | membrane protein | - |
| EN12_03070 | 640400..641191 | + | 792 | AKO74232 | hypothetical protein | - |
| EN12_03080 | 642252..642944 | + | 693 | AKO74233 | disulfide isomerase | trbB |
| EN12_03085 | 642944..645343 | + | 2400 | AKO74234 | conjugal transfer protein TraC | virb4 |
| EN12_03090 | 645336..645683 | + | 348 | AKO74235 | hypothetical protein | - |
| EN12_03095 | 645667..646179 | + | 513 | AKO74236 | peptidase | - |
| EN12_03100 | 646190..647314 | + | 1125 | AKO74237 | pilus assembly protein | traW |
| EN12_03105 | 647298..648326 | + | 1029 | AKO74238 | conjugal transfer protein TraU | traU |
| EN12_03110 | 648329..652021 | + | 3693 | AKO74239 | conjugal transfer protein TraN | traN |
| EN12_03115 | 652253..652585 | + | 333 | AKO74240 | hypothetical protein | - |
| EN12_03120 | 652616..653278 | + | 663 | AKO74241 | hypothetical protein | - |
| EN12_03125 | 653369..653971 | - | 603 | AKO74242 | hypothetical protein | - |
| EN12_03130 | 654339..654665 | + | 327 | AKO74243 | hypothetical protein | - |
| EN12_03135 | 654681..655100 | + | 420 | AKO74244 | single-stranded DNA-binding protein | - |
| EN12_03140 | 655180..655998 | + | 819 | AKO74245 | recombinase | - |
Host bacterium
| ID | 360 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEVchThd1 | GenBank | CP007653 |
| Element size | 3053204 bp | Coordinate of oriT [Strand] | 5166..5464 [+] |
| Host bacterium | Vibrio cholerae strain TSY216 | Coordinate of element | 579719..680801 |
Cargo genes
| Drug resistance gene | mph(A), tet(59), aph(6)-Id, aph(3'')-Ib, sul2, dfrA1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |