Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200399 |
| Name | oriT_ICEValZJT1 |
| Organism | Vibrio alginolyticus strain ZJ-T |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP016224 (84579..84877 [-], 299 nt) |
| oriT length | 299 nt |
| IRs (inverted repeats) | 234..242, 254..262 (AAAGCCAAA..TTTGGCTTT) 45..50, 52..57 (CAAACG..CGTTTG) 7..12, 26..31 (TTTGGC..GCCAAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 299 nt
>oriT_ICEValZJT1
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file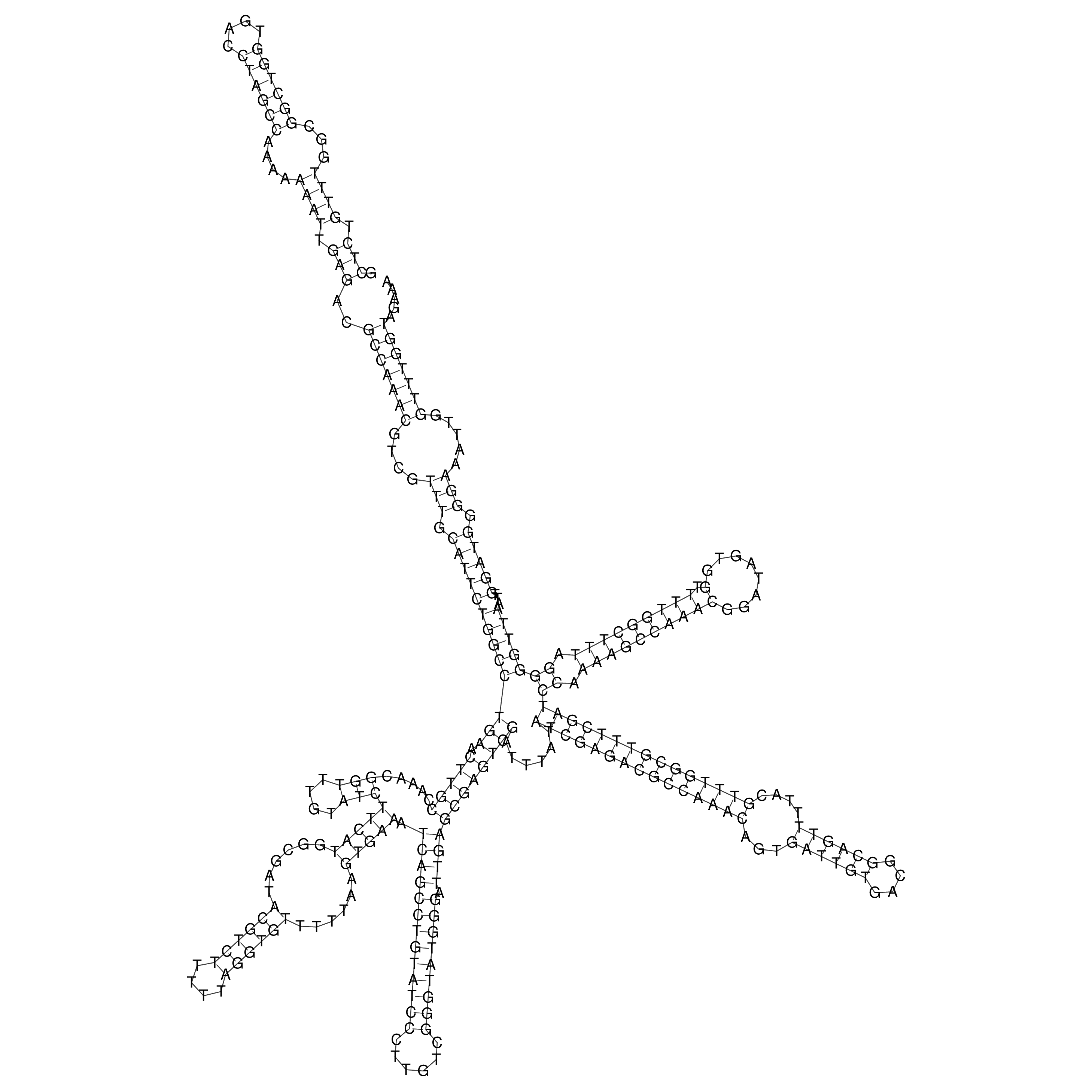
Relaxase
| ID | 14426 | GenBank | ANP65577 |
| Name | TraI_2_BAU10_11430_ICEValZJT1 |
UniProt ID | _ |
| Length | 716 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 716 a.a. Molecular weight: 79741.77 Da Isoelectric Point: 5.0396
>ANP65577.1 phosphohydrolase [Vibrio alginolyticus]
MFKNLFFQTKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQASEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKSQRVSVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWLRKSNQSTHLYLVWKSAAKDIIELLVKDKIPGIPRDPDTLADILIERGLAIKSA
SNERYESLAPEVLIKDGKPIWLPMLHISEADLLFSSNVPIGVTLFSKPEWEATQQTQAEPQSRSSEQPGL
SEASSSIELSNSAESPSTNSSEQDDERRLVSDVNHLQATENVPGDECEKPNNSYDGAISNNVNQHDAEAL
NLPESLAWLPEASSALVMVDEQILIRYPDAVRPWCAPRKLLAELSRLDWLELDPANPTRKARTVTTNDGV
QEQGLLLKVSISKGLTALIDLSNQDAESVTAIQNEEASQRPSRTETANAQAKEPATRAERKQKPIAPNAN
SSTDPKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANAHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAKR
MFKNLFFQTKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQASEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKSQRVSVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWLRKSNQSTHLYLVWKSAAKDIIELLVKDKIPGIPRDPDTLADILIERGLAIKSA
SNERYESLAPEVLIKDGKPIWLPMLHISEADLLFSSNVPIGVTLFSKPEWEATQQTQAEPQSRSSEQPGL
SEASSSIELSNSAESPSTNSSEQDDERRLVSDVNHLQATENVPGDECEKPNNSYDGAISNNVNQHDAEAL
NLPESLAWLPEASSALVMVDEQILIRYPDAVRPWCAPRKLLAELSRLDWLELDPANPTRKARTVTTNDGV
QEQGLLLKVSISKGLTALIDLSNQDAESVTAIQNEEASQRPSRTETANAQAKEPATRAERKQKPIAPNAN
SSTDPKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANAHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAKR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16949 | GenBank | ANP65563 |
| Name | t4cp2_BAU10_11350_ICEValZJT1 |
UniProt ID | _ |
| Length | 803 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 803 a.a. Molecular weight: 90516.14 Da Isoelectric Point: 5.9723
>ANP65563.1 type-IV secretion system protein TraC [Vibrio alginolyticus]
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQSIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGFDCSPP
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQSIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGFDCSPP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16950 | GenBank | ANP65576 |
| Name | t4cp2_BAU10_11425_ICEValZJT1 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67057.95 Da Isoelectric Point: 6.6444
>ANP65576.1 conjugal transfer protein [Vibrio alginolyticus]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQMG
LKGKPLAFMALSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKKTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRLPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQMG
LKGKPLAFMALSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKKTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRLPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 955247..2525324
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BAU10_04410 | 950755..953871 | + | 3117 | ANP64258 | DEAD/DEAH box helicase | - |
| BAU10_04415 | 953975..955234 | + | 1260 | ANP64259 | hypothetical protein | - |
| BAU10_04420 | 955247..956980 | + | 1734 | ANP64260 | hypothetical protein | virb4 |
| BAU10_04425 | 957098..959248 | + | 2151 | ANP64261 | phosphohydrolase | - |
| BAU10_04430 | 959296..961116 | + | 1821 | ANP64262 | conjugal transfer protein | virb4 |
| BAU10_04435 | 961126..961686 | + | 561 | ANP64263 | conjugative transfer protein | - |
| BAU10_04440 | 961673..962308 | + | 636 | ANP64264 | conjugal transfer protein | tfc7 |
| BAU10_04445 | 962446..963552 | + | 1107 | ANP64265 | cell filamentation protein Fic | - |
| BAU10_04450 | 963754..964044 | - | 291 | ANP64266 | addiction module antidote protein, HigA family | - |
| BAU10_04455 | 964277..964558 | + | 282 | ANP64267 | type IV conjugative transfer system protein TraL | traL |
| BAU10_04460 | 964555..965181 | + | 627 | ANP64268 | pilus assembly protein | traE |
| BAU10_04465 | 965165..966061 | + | 897 | ANP66442 | hypothetical protein | traK |
| BAU10_04470 | 966064..967353 | + | 1290 | ANP64269 | conjugal transfer protein TraB | traB |
| BAU10_04475 | 967350..968000 | + | 651 | ANP64270 | conjugal transfer protein TraV | traV |
| BAU10_04480 | 967997..968383 | + | 387 | ANP64271 | hypothetical protein | - |
| BAU10_04485 | 968424..968930 | - | 507 | ANP64272 | GCN5 family acetyltransferase | - |
| BAU10_04490 | 968921..969187 | - | 267 | ANP64273 | hypothetical protein | - |
| BAU10_04495 | 969556..970248 | + | 693 | ANP64274 | disulfide isomerase | trbB |
| BAU10_04500 | 970249..972651 | + | 2403 | ANP64275 | type-IV secretion system protein TraC | virb4 |
| BAU10_04505 | 972644..972991 | + | 348 | ANP64276 | plasmid-related protein | - |
| BAU10_04510 | 973068..973487 | + | 420 | ANP66443 | S26 family signal peptidase | - |
| BAU10_04515 | 973498..974622 | + | 1125 | ANP64277 | pilus assembly protein | traW |
| BAU10_04520 | 974606..975634 | + | 1029 | ANP64278 | conjugal transfer protein TraU | traU |
| BAU10_04525 | 975637..979329 | + | 3693 | ANP64279 | conjugal transfer protein TraN | traN |
| BAU10_04530 | 979560..979892 | + | 333 | ANP64280 | hypothetical protein | - |
| BAU10_04535 | 979923..980612 | + | 690 | ANP64281 | hypothetical protein | - |
| BAU10_04540 | 980590..980856 | - | 267 | ANP64282 | transposase | - |
| BAU10_04545 | 980982..981356 | + | 375 | ANP64283 | hypothetical protein | - |
| BAU10_04550 | 981402..982151 | - | 750 | Protein_872 | transposase | - |
| BAU10_04555 | 982638..982835 | - | 198 | ANP64284 | hypothetical protein | - |
| BAU10_04560 | 983143..984786 | + | 1644 | ANP64285 | BCCT transporter | - |
| BAU10_04565 | 984982..985839 | - | 858 | ANP64286 | mechanosensitive ion channel protein | - |
| BAU10_04570 | 985917..986387 | - | 471 | ANP64287 | calcium:sodium antiporter | - |
| BAU10_04575 | 986443..987378 | + | 936 | Protein_877 | transposase | - |
| BAU10_04580 | 987992..989296 | + | 1305 | ANP64288 | group II intron reverse transcriptase/maturase | - |
| BAU10_04585 | 989395..990183 | - | 789 | Protein_879 | DDE endonuclease | - |
| BAU10_04590 | 990311..991642 | - | 1332 | ANP64289 | transposase | - |
| BAU10_04595 | 991770..992372 | - | 603 | ANP64290 | hypothetical protein | - |
| BAU10_04600 | 992740..993066 | + | 327 | ANP64291 | plasmid-related protein | - |
| BAU10_04605 | 993082..993501 | + | 420 | ANP64292 | single-stranded DNA-binding protein | - |
| BAU10_04610 | 993581..994399 | + | 819 | ANP64293 | phage recombination protein Bet | - |
| BAU10_04615 | 994686..995702 | + | 1017 | ANP64294 | endonuclease | - |
| BAU10_04620 | 995912..996871 | + | 960 | ANP64295 | ATPase | - |
| BAU10_04625 | 996871..997638 | + | 768 | ANP64296 | hypothetical protein | - |
| BAU10_04630 | 997737..998690 | + | 954 | ANP64297 | cobalamin biosynthesis protein | - |
| BAU10_04635 | 998752..999192 | + | 441 | ANP64298 | hypothetical protein | - |
| BAU10_04640 | 999262..1000917 | + | 1656 | ANP64299 | hypothetical protein | - |
| BAU10_04645 | 1001001..1001498 | + | 498 | ANP64300 | DNA repair protein RadC | - |
| BAU10_04650 | 1001498..1001839 | + | 342 | ANP64301 | plasmid-related protein | - |
| BAU10_04655 | 1001930..1003003 | + | 1074 | ANP64302 | DNA primase | - |
| BAU10_04660 | 1003092..1003799 | + | 708 | ANP64303 | hypothetical protein | - |
| BAU10_04665 | 1003956..1004288 | + | 333 | ANP64304 | hypothetical protein | - |
| BAU10_04670 | 1004385..1005737 | - | 1353 | ANP64305 | hypothetical protein | - |
| BAU10_04675 | 1006253..1007047 | + | 795 | ANP64306 | hypothetical protein | - |
| BAU10_04680 | 1007218..1007691 | - | 474 | Protein_898 | hypothetical protein | - |
| BAU10_04685 | 1007805..1008236 | - | 432 | ANP64307 | hypothetical protein | - |
| BAU10_04690 | 1008233..1008706 | - | 474 | ANP64308 | hypothetical protein | - |
| BAU10_04695 | 1008852..1009031 | + | 180 | ANP64309 | transcriptional regulator | - |
| BAU10_04700 | 1009208..1009477 | + | 270 | ANP64310 | hypothetical protein | - |
| BAU10_04705 | 1009558..1010238 | - | 681 | ANP64311 | two-component response regulator DpiA | - |
| BAU10_04710 | 1010225..1011880 | - | 1656 | ANP64312 | ATPase | - |
| BAU10_04715 | 1011993..1013294 | - | 1302 | ANP64313 | oxaloacetate decarboxylase subunit beta | - |
| BAU10_04720 | 1013310..1015121 | - | 1812 | ANP64314 | oxaloacetate decarboxylase subunit alpha | - |
| BAU10_04725 | 1015133..1015384 | - | 252 | ANP64315 | oxaloacetate decarboxylase | - |
| BAU10_04730 | 1015453..1016796 | - | 1344 | ANP66444 | citrate:sodium symporter | - |
| BAU10_04735 | 1017067..1018119 | + | 1053 | ANP66445 | [citrate (pro-3S)-lyase] ligase | - |
| BAU10_04740 | 1018161..1018457 | + | 297 | ANP64316 | citrate lyase acyl carrier protein | - |
| BAU10_04745 | 1018454..1019329 | + | 876 | ANP64317 | citrate (pro-3S)-lyase subunit beta | - |
| BAU10_04750 | 1019384..1020856 | + | 1473 | ANP66446 | citrate lyase subunit alpha | - |
| BAU10_04755 | 1020853..1021389 | + | 537 | ANP64318 | holo-ACP synthase CitX | - |
| BAU10_04760 | 1021370..1022329 | + | 960 | ANP64319 | triphosphoribosyl-dephospho-CoA synthase CitG | - |
| BAU10_04765 | 1022348..1023175 | + | 828 | ANP64320 | hypothetical protein | - |
| BAU10_04770 | 1023592..1025205 | + | 1614 | Protein_916 | transposase | - |
| BAU10_04775 | 1025226..1025738 | - | 513 | Protein_917 | transposase | - |
| BAU10_04780 | 1026108..1027052 | + | 945 | ANP64321 | pilus assembly protein | - |
| BAU10_04785 | 1027055..1028443 | + | 1389 | ANP64322 | conjugal transfer protein TraH | - |
| BAU10_04790 | 1028447..1032016 | + | 3570 | ANP64323 | conjugal transfer protein TraG | - |
| BAU10_04795 | 1032049..1032480 | - | 432 | ANP64324 | hypothetical protein | - |
| BAU10_04800 | 1032538..1033071 | - | 534 | ANP64325 | transcriptional regulator | - |
| BAU10_04805 | 1033068..1033367 | - | 300 | ANP64326 | transcriptional regulator | - |
| BAU10_04810 | 1033364..1033912 | - | 549 | ANP64327 | transglycosylase | - |
| BAU10_04815 | 1033899..1034561 | - | 663 | ANP64328 | hypothetical protein | - |
| BAU10_04820 | 1034548..1035417 | - | 870 | ANP64329 | hypothetical protein | - |
| BAU10_04825 | 1035473..1035724 | - | 252 | ANP64330 | Cro/Cl family transcriptional regulator | - |
| BAU10_04830 | 1035842..1036489 | + | 648 | ANP64331 | transcriptional regulator | - |
| BAU10_04835 | 1037396..1038388 | - | 993 | ANP64332 | cytochrome BD ubiquinol oxidase subunit II | - |
| BAU10_04840 | 1038391..1039722 | - | 1332 | ANP64333 | cytochrome D ubiquinol oxidase subunit I | - |
| BAU10_04845 | 1040538..1040786 | + | 249 | ANP64334 | hypothetical protein | - |
| BAU10_04850 | 1040792..1041781 | - | 990 | ANP64335 | hypothetical protein | - |
| BAU10_04855 | 1042556..1044193 | + | 1638 | ANP64336 | chemotaxis protein | - |
| BAU10_04860 | 1044659..1045798 | + | 1140 | ANP64337 | efflux transporter periplasmic adaptor subunit | - |
| BAU10_04865 | 1045801..1048962 | + | 3162 | ANP64338 | multidrug efflux RND transporter permease | - |
| BAU10_04870 | 1049150..1049605 | + | 456 | ANP64339 | transcriptional regulator | - |
| BAU10_04875 | 1049650..1050606 | - | 957 | ANP64340 | transporter | - |
| BAU10_04880 | 1051208..1052230 | + | 1023 | ANP64341 | tryptophan--tRNA ligase | - |
| BAU10_04885 | 1052315..1053271 | - | 957 | ANP64342 | ATP-dependent dsDNA exonuclease | - |
| BAU10_04890 | 1053268..1054596 | - | 1329 | ANP64343 | phospholipase | - |
| BAU10_04895 | 1054888..1055154 | + | 267 | ANP64344 | hypothetical protein | - |
| BAU10_04900 | 1055309..1056937 | + | 1629 | ANP64345 | chemotaxis protein | - |
| BAU10_04905 | 1057063..1057833 | - | 771 | ANP64346 | tRNA hydroxylase | - |
| BAU10_04910 | 1057931..1058695 | - | 765 | ANP64347 | DNA repair protein | - |
| BAU10_04915 | 1059009..1059983 | + | 975 | ANP64348 | transcriptional regulator CysB | - |
| BAU10_04920 | 1060037..1061233 | + | 1197 | ANP64349 | SAM-dependent methyltransferase | - |
| BAU10_04925 | 1061327..1062451 | - | 1125 | ANP64350 | alanine dehydrogenase | - |
| BAU10_04930 | 1062608..1063102 | + | 495 | ANP64351 | leucine-responsive transcriptional regulator | - |
| BAU10_04935 | 1063255..1066302 | + | 3048 | ANP64352 | cell division protein FtsK | - |
| BAU10_04940 | 1066364..1066990 | + | 627 | ANP64353 | outer membrane lipoprotein carrier protein LolA | - |
| BAU10_04945 | 1066997..1068346 | + | 1350 | ANP66447 | recombination factor protein RarA | - |
| BAU10_04950 | 1068438..1069745 | + | 1308 | ANP64354 | serine--tRNA ligase | - |
| BAU10_04955 | 1069942..1070208 | + | 267 | ANP64355 | hypothetical protein | - |
| BAU10_04960 | 1070278..1071558 | - | 1281 | ANP64356 | adenosylmethionine--8-amino-7-oxononanoate transaminase | - |
| BAU10_04965 | 1071689..1072741 | + | 1053 | ANP64357 | biotin synthase BioB | - |
| BAU10_04970 | 1072728..1073879 | + | 1152 | ANP64358 | 8-amino-7-oxononanoate synthase | - |
| BAU10_04975 | 1073879..1074700 | + | 822 | ANP64359 | malonyl-[acyl-carrier protein] O-methyltransferase BioC | - |
| BAU10_04980 | 1074697..1075383 | + | 687 | ANP64360 | dethiobiotin synthase | - |
| BAU10_04985 | 1076142..1077011 | + | 870 | ANP64361 | zinc metalloprotease HtpX | - |
| BAU10_04990 | 1077221..1077640 | + | 420 | ANP64362 | transcriptional regulator | - |
| BAU10_04995 | 1077637..1078362 | + | 726 | ANP64363 | short-chain dehydrogenase | - |
| BAU10_05000 | 1078375..1079685 | + | 1311 | ANP64364 | FAD-dependent oxidoreductase | - |
| BAU10_05005 | 1079687..1080505 | + | 819 | ANP64365 | chromosome partitioning protein ParA | - |
| BAU10_05010 | 1080499..1081755 | + | 1257 | ANP64366 | cyclopropane-fatty-acyl-phospholipid synthase | - |
| BAU10_05015 | 1081762..1082274 | + | 513 | ANP64367 | zinc ABC transporter permease | - |
| BAU10_05020 | 1082234..1082827 | + | 594 | ANP64368 | hypothetical protein | - |
| BAU10_05025 | 1082839..1083360 | + | 522 | ANP66448 | hypothetical protein | - |
| BAU10_05030 | 1083494..1084864 | - | 1371 | ANP64369 | adenylosuccinate lyase | - |
| BAU10_05035 | 1085018..1085635 | - | 618 | ANP64370 | lysogenization protein HflD | - |
| BAU10_05040 | 1085646..1086770 | - | 1125 | ANP64371 | tRNA 2-thiouridine(34) synthase MnmA | - |
| BAU10_05045 | 1086959..1087228 | + | 270 | ANP64372 | hypothetical protein | - |
| BAU10_05050 | 1087346..1088650 | + | 1305 | ANP64373 | inosine/guanosine kinase | - |
| BAU10_05055 | 1088778..1089185 | - | 408 | ANP64374 | transcriptional regulator | - |
| BAU10_05060 | 1090060..1091661 | + | 1602 | ANP64375 | Na+/H+ antiporter | - |
| BAU10_05065 | 1091663..1091854 | - | 192 | ANP64376 | hypothetical protein | - |
| BAU10_05070 | 1092033..1092929 | + | 897 | ANP64377 | ATP phosphoribosyltransferase | - |
| BAU10_05075 | 1092935..1094227 | + | 1293 | ANP64378 | histidinol dehydrogenase | - |
| BAU10_05080 | 1094246..1095286 | + | 1041 | ANP64379 | histidinol-phosphate transaminase | - |
| BAU10_05085 | 1095356..1096429 | + | 1074 | ANP64380 | bifunctional imidazole glycerol-phosphate dehydratase/histidinol phosphatase | - |
| BAU10_05090 | 1096456..1097070 | + | 615 | ANP64381 | imidazole glycerol phosphate synthase, glutamine amidotransferase subunit | - |
| BAU10_05095 | 1097083..1097820 | + | 738 | ANP64382 | 1-(5-phosphoribosyl)-5-[(5- phosphoribosylamino)methylideneamino]imidazole-4- carboxamide isomerase | - |
| BAU10_05100 | 1097802..1098575 | + | 774 | ANP64383 | imidazole glycerol phosphate synthase subunit HisF | - |
| BAU10_05105 | 1098572..1099207 | + | 636 | ANP64384 | bifunctional phosphoribosyl-AMP cyclohydrolase/phosphoribosyl-ATP diphosphatase | - |
| BAU10_05110 | 1099488..1100003 | - | 516 | ANP64385 | hypothetical protein | - |
| BAU10_05115 | 1100084..1101298 | - | 1215 | ANP64386 | histidine kinase | - |
| BAU10_05120 | 1101397..1102134 | - | 738 | ANP64387 | UDP-2,3-diacylglucosamine diphosphatase | - |
| BAU10_05125 | 1102195..1102689 | - | 495 | ANP64388 | peptidylprolyl isomerase | - |
| BAU10_05130 | 1102892..1104274 | + | 1383 | ANP64389 | cysteine--tRNA ligase | - |
| BAU10_05135 | 1104362..1104940 | + | 579 | ANP64390 | thymidine kinase | - |
| BAU10_05140 | 1105001..1105639 | - | 639 | ANP64391 | hypothetical protein | - |
| BAU10_05145 | 1105636..1106463 | - | 828 | ANP64392 | NAD(P)-dependent oxidoreductase | - |
| BAU10_05150 | 1106482..1107360 | - | 879 | ANP64393 | LysR family transcriptional regulator | - |
| BAU10_05155 | 1107450..1107896 | + | 447 | ANP64394 | hypothetical protein | - |
| BAU10_05160 | 1107940..1108173 | - | 234 | ANP64395 | TIGR02647 family protein | - |
| BAU10_05165 | 1108473..1109333 | + | 861 | ANP64396 | formate transporter FocA | - |
| BAU10_05170 | 1109911..1110723 | - | 813 | ANP64397 | ABC transporter permease | - |
| BAU10_05175 | 1111141..1111425 | - | 285 | ANP64398 | hypothetical protein | - |
| BAU10_05180 | 1111416..1112600 | + | 1185 | ANP64399 | nitrate reductase | - |
| BAU10_05185 | 1112627..1115089 | + | 2463 | ANP64400 | trimethylamine N-oxide reductase I catalytic subunit | - |
| BAU10_05190 | 1115288..1117027 | + | 1740 | ANP64401 | alpha-galactosidase | - |
| BAU10_05195 | 1117106..1118320 | - | 1215 | ANP64402 | hypothetical protein | - |
| BAU10_05200 | 1118507..1119412 | + | 906 | ANP64403 | manganese-dependent inorganic pyrophosphatase | - |
| BAU10_05205 | 1119481..1119870 | + | 390 | ANP64404 | hypothetical protein | - |
| BAU10_05210 | 1119952..1120743 | - | 792 | ANP64405 | peptide ABC transporter ATP-binding protein | - |
| BAU10_05215 | 1120740..1121735 | - | 996 | ANP64406 | peptide ABC transporter ATP-binding protein | - |
| BAU10_05220 | 1121735..1122622 | - | 888 | ANP64407 | peptide ABC transporter permease | - |
| BAU10_05225 | 1122609..1123571 | - | 963 | ANP64408 | peptide ABC transporter permease | - |
| BAU10_05230 | 1123571..1125190 | - | 1620 | ANP64409 | ABC transporter substrate-binding protein | - |
| BAU10_05235 | 1125396..1126403 | - | 1008 | ANP64410 | phage shock protein operon transcriptional activator | - |
| BAU10_05240 | 1126636..1127307 | + | 672 | ANP64411 | phage shock protein PspA | - |
| BAU10_05245 | 1127322..1127555 | + | 234 | ANP64412 | phage shock protein B | - |
| BAU10_05250 | 1127548..1127937 | + | 390 | ANP64413 | phage shock protein C | - |
| BAU10_05255 | 1128099..1129130 | + | 1032 | ANP64414 | efflux transporter periplasmic adaptor subunit | - |
| BAU10_05260 | 1129132..1130229 | + | 1098 | ANP64415 | efflux transporter periplasmic adaptor subunit | - |
| BAU10_05265 | 1130226..1133336 | + | 3111 | ANP64416 | multidrug transporter AcrB | - |
| BAU10_05270 | 1133409..1133975 | + | 567 | ANP64417 | DNA-3-methyladenine glycosidase | - |
| BAU10_05275 | 1134000..1134860 | - | 861 | ANP64418 | lipase chaperone | - |
| BAU10_05280 | 1134873..1135778 | - | 906 | ANP64419 | lipase | - |
| BAU10_05285 | 1136027..1137232 | - | 1206 | ANP64420 | cystathionine beta-lyase | - |
| BAU10_05290 | 1137444..1138898 | - | 1455 | ANP64421 | cardiolipin synthase | - |
| BAU10_05295 | 1139329..1140699 | + | 1371 | ANP66449 | Na+-dependent transporter | - |
| BAU10_05300 | 1140782..1141474 | + | 693 | ANP64422 | RNA pseudouridine synthase | - |
| BAU10_05305 | 1141506..1142447 | + | 942 | ANP64423 | methyltransferase | - |
| BAU10_05310 | 1142661..1143716 | - | 1056 | ANP64424 | hybrid-cluster NAD(P)-dependent oxidoreductase | - |
| BAU10_05315 | 1143798..1145459 | - | 1662 | ANP64425 | hydroxylamine reductase | - |
| BAU10_05320 | 1145634..1147163 | - | 1530 | ANP64426 | nitric oxide reductase transcription regulator | - |
| BAU10_05325 | 1147366..1147845 | - | 480 | ANP64427 | hypothetical protein | - |
| BAU10_05330 | 1147969..1148580 | - | 612 | ANP64428 | hypothetical protein | - |
| BAU10_05335 | 1148573..1149220 | - | 648 | ANP64429 | ABC transporter ATP-binding protein | - |
| BAU10_05340 | 1149186..1150895 | - | 1710 | ANP64430 | thiamine ABC transporter permease | - |
| BAU10_05345 | 1150905..1152065 | - | 1161 | ANP64431 | hypothetical protein | - |
| BAU10_05350 | 1152501..1152947 | - | 447 | ANP64432 | heat-shock protein HslJ | - |
| BAU10_05355 | 1153132..1153380 | + | 249 | ANP64433 | Fe-S protein | - |
| BAU10_05360 | 1153516..1153737 | - | 222 | ANP64434 | hypothetical protein | - |
| BAU10_05365 | 1154153..1155841 | - | 1689 | ANP64435 | bifunctional UDP-sugar hydrolase/5'-nucleotidase | - |
| BAU10_05370 | 1156594..1157109 | + | 516 | ANP64436 | GNAT family N-acetyltransferase | - |
| BAU10_05375 | 1157257..1157535 | - | 279 | ANP64437 | 50S ribosomal protein L25 | - |
| BAU10_05380 | 1157819..1159168 | - | 1350 | ANP64438 | histidine kinase | - |
| BAU10_05385 | 1159158..1159817 | - | 660 | ANP64439 | DNA-binding response regulator | - |
| BAU10_05390 | 1159894..1160319 | - | 426 | ANP64440 | hypothetical protein | - |
| BAU10_05395 | 1160415..1162166 | - | 1752 | ANP64441 | ATP-dependent helicase | - |
| BAU10_05400 | 1162288..1162989 | + | 702 | ANP64442 | 16S rRNA pseudouridine(516) synthase | - |
| BAU10_05405 | 1163109..1163390 | - | 282 | ANP64443 | hypothetical protein | - |
| BAU10_05410 | 1163426..1164628 | + | 1203 | ANP64444 | Bcr/CflA family drug resistance efflux transporter | - |
| BAU10_05415 | 1164628..1165290 | + | 663 | ANP64445 | alpha-acetolactate decarboxylase | - |
| BAU10_05420 | 1165321..1166772 | - | 1452 | ANP64446 | NAD(P)-dependent oxidoreductase | - |
| BAU10_05425 | 1166867..1168243 | + | 1377 | ANP64447 | MATE family efflux transporter | - |
| BAU10_05430 | 1168431..1169102 | + | 672 | ANP64448 | ABC transporter ATP-binding protein | - |
| BAU10_05435 | 1169083..1171536 | + | 2454 | ANP64449 | ABC transporter permease | - |
| BAU10_05440 | 1171536..1172660 | + | 1125 | ANP64450 | ABC transporter | - |
| BAU10_05445 | 1172991..1174142 | + | 1152 | ANP64451 | Na(+)/H(+) antiporter NhaA | - |
| BAU10_05450 | 1174860..1175495 | + | 636 | ANP64452 | TetR family transcriptional regulator | - |
| BAU10_05455 | 1175567..1177849 | + | 2283 | ANP64453 | acyl-CoA dehydrogenase | - |
| BAU10_05460 | 1178024..1179226 | + | 1203 | ANP64454 | trans-2-enoyl-CoA reductase | - |
| BAU10_05465 | 1179305..1180987 | - | 1683 | ANP64455 | transporter | - |
| BAU10_05470 | 1181163..1181429 | - | 267 | ANP64456 | glutaredoxin | - |
| BAU10_05475 | 1181807..1183051 | + | 1245 | ANP64457 | alcohol dehydrogenase | - |
| BAU10_05480 | 1183128..1183982 | - | 855 | ANP64458 | transcriptional regulator HexR | - |
| BAU10_05485 | 1184294..1185940 | - | 1647 | ANP64459 | glutamate decarboxylase | - |
| BAU10_05490 | 1186152..1186664 | - | 513 | ANP64460 | hypothetical protein | - |
| BAU10_05495 | 1186873..1187778 | - | 906 | ANP64461 | hypothetical protein | - |
| BAU10_05500 | 1187925..1188845 | - | 921 | ANP64462 | histidine kinase | - |
| BAU10_05505 | 1189215..1189802 | + | 588 | ANP64463 | DNA transformation protein | - |
| BAU10_05510 | 1189995..1190543 | - | 549 | ANP64464 | hypothetical protein | - |
| BAU10_05515 | 1190835..1191332 | - | 498 | ANP64465 | hypothetical protein | - |
| BAU10_05520 | 1191329..1193269 | - | 1941 | ANP66450 | ATPase | - |
| BAU10_05525 | 1193456..1194724 | + | 1269 | ANP64466 | hypothetical protein | - |
| BAU10_05530 | 1194862..1195956 | + | 1095 | ANP64467 | phosphoserine transaminase | - |
| BAU10_05535 | 1196047..1197378 | + | 1332 | ANP64468 | D-serine ammonia-lyase | - |
| BAU10_05540 | 1197477..1199198 | - | 1722 | ANP64469 | cysteine/glutathione ABC transporter ATP-binding protein/permease CydC | - |
| BAU10_05545 | 1199191..1200978 | - | 1788 | ANP64470 | thiol reductant ABC exporter subunit CydD | - |
| BAU10_05550 | 1201082..1202041 | - | 960 | ANP64471 | thioredoxin-disulfide reductase | - |
| BAU10_05555 | 1202401..1203057 | + | 657 | ANP64472 | agglutination protein | - |
| BAU10_05560 | 1203041..1204276 | + | 1236 | ANP64473 | cysteine desulfurase-like protein | - |
| BAU10_05565 | 1204367..1205203 | - | 837 | ANP64474 | hypothetical protein | - |
| BAU10_05570 | 1205540..1206877 | + | 1338 | ANP66451 | diguanylate cyclase | - |
| BAU10_05575 | 1207021..1208391 | - | 1371 | ANP64475 | Anion transporter | - |
| BAU10_05580 | 1208611..1209345 | - | 735 | ANP64476 | SanA protein | - |
| BAU10_05585 | 1209500..1211188 | - | 1689 | ANP64477 | NAD-dependent malic enzyme | - |
| BAU10_05590 | 1211402..1213756 | + | 2355 | ANP66452 | DEAD/DEAH box helicase | - |
| BAU10_05595 | 1214045..1214422 | + | 378 | ANP64478 | transporter | - |
| BAU10_05600 | 1214752..1216425 | + | 1674 | ANP64479 | alkaline phosphatase | - |
| BAU10_05605 | 1216504..1217607 | - | 1104 | ANP64480 | phosphoribosylaminoimidazolesuccinocarboxamide synthase | - |
| BAU10_05610 | 1217811..1218203 | - | 393 | ANP64481 | hypothetical protein | - |
| BAU10_05615 | 1218615..1218797 | - | 183 | ANP64482 | hypothetical protein | - |
| BAU10_05620 | 1218975..1219949 | - | 975 | ANP64483 | molecular chaperone DnaJ | - |
| BAU10_05625 | 1220026..1220694 | - | 669 | ANP64484 | hypothetical protein | - |
| BAU10_05630 | 1220903..1221598 | - | 696 | ANP64485 | transcriptional regulator | - |
| BAU10_05635 | 1221807..1222487 | - | 681 | ANP64486 | chemotaxis protein CheX | - |
| BAU10_05640 | 1222749..1225784 | + | 3036 | ANP64487 | hypothetical protein | - |
| BAU10_05645 | 1225858..1226319 | + | 462 | ANP64488 | hypothetical protein | - |
| BAU10_05650 | 1226430..1227155 | - | 726 | ANP64489 | hypothetical protein | - |
| BAU10_05655 | 1227491..1229026 | - | 1536 | ANP64490 | histidine ammonia-lyase | - |
| BAU10_05660 | 1229038..1230735 | - | 1698 | ANP66453 | urocanate hydratase | - |
| BAU10_05665 | 1230809..1231819 | - | 1011 | ANP64491 | formimidoylglutamase | - |
| BAU10_05670 | 1231821..1233071 | - | 1251 | ANP64492 | imidazolonepropionase | - |
| BAU10_05675 | 1233201..1233908 | - | 708 | ANP64493 | histidine utilization repressor | - |
| BAU10_05680 | 1234157..1234900 | + | 744 | ANP64494 | sporulation control protein Spo0M | - |
| BAU10_05685 | 1234907..1235884 | - | 978 | ANP64495 | hypothetical protein | - |
| BAU10_05690 | 1236288..1238216 | + | 1929 | ANP64496 | threonine--tRNA ligase | - |
| BAU10_05695 | 1238382..1238771 | + | 390 | ANP64497 | translation initiation factor IF-3 | - |
| BAU10_05700 | 1238893..1239087 | + | 195 | ANP64498 | 50S ribosomal protein L35 | - |
| BAU10_05705 | 1239128..1239481 | + | 354 | ANP64499 | 50S ribosomal protein L20 | - |
| BAU10_05710 | 1239671..1239988 | + | 318 | ANP64500 | hypothetical protein | - |
| BAU10_05715 | 1240355..1241164 | + | 810 | ANP64501 | hypothetical protein | - |
| BAU10_05720 | 1241157..1241342 | - | 186 | ANP64502 | hypothetical protein | - |
| BAU10_05725 | 1241613..1242401 | + | 789 | ANP64503 | TetR family transcriptional regulator | - |
| BAU10_05730 | 1242651..1245029 | + | 2379 | ANP64504 | RND transporter | - |
| BAU10_05735 | 1245048..1245839 | + | 792 | ANP64505 | outer membrane lipoprotein-sorting protein | - |
| BAU10_05740 | 1245850..1247025 | + | 1176 | ANP64506 | hypothetical protein | - |
| BAU10_05745 | 1247217..1248200 | + | 984 | ANP64507 | phenylalanine--tRNA ligase subunit alpha | - |
| BAU10_05750 | 1248219..1250624 | + | 2406 | ANP64508 | phenylalanine--tRNA ligase subunit beta | - |
| BAU10_05755 | 1250838..1251719 | - | 882 | ANP64509 | hypothetical protein | - |
| BAU10_05760 | 1252254..1252550 | + | 297 | ANP64510 | integration host factor subunit alpha | - |
| BAU10_05765 | 1252672..1253451 | - | 780 | ANP64511 | phosphoribosylglycinamide formyltransferase | - |
| BAU10_05770 | 1253767..1254417 | + | 651 | ANP66454 | thiopurine S-methyltransferase | - |
| BAU10_05775 | 1254490..1255665 | - | 1176 | ANP64512 | phosphoribosylglycinamide formyltransferase 2 | - |
| BAU10_05780 | 1255809..1256696 | - | 888 | ANP64513 | cytidine deaminase | - |
| BAU10_05785 | 1257128..1257805 | - | 678 | ANP64514 | CidB/LrgB family autolysis modulator | - |
| BAU10_05790 | 1257807..1258181 | - | 375 | ANP64515 | hypothetical protein | - |
| BAU10_05795 | 1258237..1259658 | - | 1422 | ANP64516 | exodeoxyribonuclease I | - |
| BAU10_05800 | 1259816..1261189 | - | 1374 | ANP64517 | L-cystine transporter tcyP | - |
| BAU10_05805 | 1261390..1262079 | - | 690 | ANP64518 | nucleoside-diphosphate sugar epimerase | - |
| BAU10_05810 | 1262329..1263372 | + | 1044 | ANP64519 | nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | - |
| BAU10_05815 | 1263372..1264178 | + | 807 | ANP64520 | adenosylcobinamide-GDP ribazoletransferase | - |
| BAU10_05820 | 1264175..1264708 | + | 534 | ANP64521 | bifunctional adenosylcobinamide kinase/adenosylcobinamide-phosphate guanylyltransferase | - |
| BAU10_05825 | 1264720..1265334 | + | 615 | ANP64522 | alpha-ribazole phosphatase | - |
| BAU10_05830 | 1265463..1267259 | + | 1797 | ANP64523 | oligoendopeptidase F | - |
| BAU10_05835 | 1267534..1268565 | + | 1032 | ANP64524 | succinylglutamate desuccinylase | - |
| BAU10_05840 | 1268648..1269643 | + | 996 | ANP64525 | vitamin B12 ABC transporter permease BtuC | - |
| BAU10_05845 | 1269633..1270397 | + | 765 | ANP64526 | vitamin B12 ABC transporter ATP-binding protein BtuD | - |
| BAU10_05850 | 1270467..1271042 | - | 576 | ANP64527 | SAM-dependent methyltransferase | - |
| BAU10_05855 | 1271152..1271724 | - | 573 | ANP64528 | hypothetical protein | - |
| BAU10_05860 | 1271815..1273038 | - | 1224 | ANP64529 | MFS transporter permease | - |
| BAU10_05865 | 1273145..1274014 | + | 870 | ANP64530 | LysR family transcriptional regulator | - |
| BAU10_05870 | 1274042..1274272 | - | 231 | ANP64531 | NrdH-redoxin | - |
| BAU10_05875 | 1274411..1274926 | + | 516 | ANP64532 | metal-dependent hydrolase | - |
| BAU10_05880 | 1274913..1276040 | + | 1128 | ANP64533 | 23S rRNA (adenine(1618)-N(6))-methyltransferase | - |
| BAU10_05885 | 1276113..1276493 | - | 381 | ANP66455 | hypothetical protein | - |
| BAU10_05890 | 1276504..1278099 | - | 1596 | ANP64534 | peptidase S41 | - |
| BAU10_05895 | 1278458..1279222 | - | 765 | ANP64535 | transcriptional regulator | - |
| BAU10_05900 | 1279253..1279927 | - | 675 | ANP64536 | GntR family transcriptional regulator | - |
| BAU10_05905 | 1280080..1281612 | - | 1533 | ANP64537 | aldehyde dehydrogenase | - |
| BAU10_05910 | 1281634..1282635 | - | 1002 | ANP64538 | hydroxyproline-2-epimerase | - |
| BAU10_05915 | 1282745..1284001 | + | 1257 | ANP64539 | amino acid dehydrogenase | - |
| BAU10_05920 | 1284125..1285198 | + | 1074 | ANP64540 | spermidine/putrescine ABC transporter substrate-binding protein | - |
| BAU10_05925 | 1285285..1286259 | + | 975 | ANP64541 | ornithine cyclodeaminase | - |
| BAU10_05930 | 1286316..1287230 | + | 915 | ANP64542 | dihydrodipicolinate synthase family protein | - |
| BAU10_05935 | 1287304..1288425 | + | 1122 | ANP64543 | spermidine/putrescine ABC transporter ATP-binding protein | - |
| BAU10_05940 | 1288422..1289702 | + | 1281 | ANP64544 | ABC transporter permease | - |
| BAU10_05945 | 1289705..1290553 | + | 849 | ANP64545 | polyamine ABC transporter permease | - |
| BAU10_05950 | 1290674..1291108 | - | 435 | ANP64546 | hypothetical protein | - |
| BAU10_05955 | 1291411..1293855 | + | 2445 | ANP64547 | collagenase | - |
| BAU10_05960 | 1294016..1295812 | - | 1797 | ANP64548 | Xaa-Pro aminopeptidase | - |
| BAU10_05965 | 1295812..1296804 | - | 993 | ANP64549 | oligopeptide ABC transporter ATP-binding protein OppF | - |
| BAU10_05970 | 1296814..1297797 | - | 984 | ANP64550 | oligopeptide transporter ATP-binding component | - |
| BAU10_05975 | 1297808..1298740 | - | 933 | ANP64551 | peptide ABC transporter permease | - |
| BAU10_05980 | 1298750..1299670 | - | 921 | ANP64552 | oligopeptide transporter permease | - |
| BAU10_05985 | 1299815..1301425 | - | 1611 | ANP64553 | peptide ABC transporter substrate-binding protein | - |
| BAU10_05990 | 1301690..1302796 | - | 1107 | ANP64554 | peptidase M20 | - |
| BAU10_05995 | 1303335..1304408 | + | 1074 | ANP64555 | 4-hydroxyphenylpyruvate dioxygenase | - |
| BAU10_06000 | 1304401..1305531 | + | 1131 | ANP64556 | homogentisate 1,2-dioxygenase | - |
| BAU10_06005 | 1305541..1306578 | + | 1038 | ANP64557 | 2-keto-4-pentenoate hydratase | - |
| BAU10_06010 | 1306600..1307268 | + | 669 | ANP64558 | maleylacetoacetate isomerase | - |
| BAU10_06015 | 1307464..1308123 | + | 660 | ANP64559 | hypothetical protein | - |
| BAU10_06020 | 1308243..1308581 | + | 339 | ANP64560 | hypothetical protein | - |
| BAU10_06035 | 1309206..1309445 | - | 240 | ANP64561 | hypothetical protein | - |
| BAU10_06040 | 1309566..1310204 | - | 639 | ANP64562 | thiamine biosynthesis protein ThiJ | - |
| BAU10_06045 | 1310370..1310768 | - | 399 | ANP64563 | glyoxalase | - |
| BAU10_06050 | 1310974..1311612 | - | 639 | ANP64564 | hypothetical protein | - |
| BAU10_06055 | 1311739..1312188 | - | 450 | ANP64565 | hypothetical protein | - |
| BAU10_06060 | 1312352..1312741 | - | 390 | ANP64566 | hypothetical protein | - |
| BAU10_06065 | 1313146..1313397 | - | 252 | ANP64567 | TIGR03643 family protein | - |
| BAU10_06070 | 1313496..1313996 | - | 501 | ANP64568 | histone acetyltransferase | - |
| BAU10_06075 | 1314147..1314527 | - | 381 | ANP64569 | hypothetical protein | - |
| BAU10_06080 | 1314681..1315085 | - | 405 | ANP64570 | hypothetical protein | - |
| BAU10_06085 | 1315227..1315559 | - | 333 | ANP64571 | hypothetical protein | - |
| BAU10_06090 | 1315624..1316091 | - | 468 | ANP64572 | hypothetical protein | - |
| BAU10_06095 | 1316508..1317191 | - | 684 | ANP64573 | hypothetical protein | - |
| BAU10_06100 | 1317441..1318916 | - | 1476 | ANP64574 | sodium:proton antiporter | - |
| BAU10_06105 | 1318935..1320278 | - | 1344 | ANP64575 | gamma-glutamylputrescine synthetase | - |
| BAU10_06110 | 1320619..1321365 | + | 747 | ANP64576 | gamma-glutamyl-gamma-aminobutyrate hydrolase | - |
| BAU10_06115 | 1321384..1321941 | + | 558 | ANP64577 | XRE family transcriptional regulator | - |
| BAU10_06120 | 1321977..1323461 | + | 1485 | ANP64578 | aldehyde dehydrogenase PuuC | - |
| BAU10_06125 | 1323499..1324776 | + | 1278 | ANP64579 | gamma-glutamylputrescine oxidoreductase | - |
| BAU10_06130 | 1324794..1325660 | + | 867 | ANP64580 | N-carbamoylputrescine amidase | - |
| BAU10_06135 | 1325673..1326755 | + | 1083 | ANP64581 | agmatine deiminase | - |
| BAU10_06140 | 1326810..1328237 | + | 1428 | ANP64582 | succinate-semialdehyde dehydrogenase (NADP(+)) | - |
| BAU10_06145 | 1328252..1329523 | + | 1272 | ANP64583 | 4-aminobutyrate transaminase | - |
| BAU10_06150 | 1329870..1331366 | + | 1497 | ANP64584 | diguanylate phosphodiesterase | - |
| BAU10_06155 | 1331498..1332385 | - | 888 | ANP64585 | multidrug transporter | - |
| BAU10_06160 | 1332496..1332978 | + | 483 | ANP64586 | transcriptional regulator | - |
| BAU10_06165 | 1333002..1333355 | + | 354 | ANP64587 | hypothetical protein | - |
| BAU10_06170 | 1333475..1333909 | + | 435 | ANP64588 | hypothetical protein | - |
| BAU10_06175 | 1334112..1334423 | + | 312 | ANP64589 | hypothetical protein | - |
| BAU10_06180 | 1334570..1335661 | - | 1092 | ANP64590 | sulfate ABC transporter permease | - |
| BAU10_06185 | 1335663..1336139 | - | 477 | ANP64591 | hypothetical protein | - |
| BAU10_06190 | 1336149..1339319 | - | 3171 | ANP66456 | hybrid sensor histidine kinase/response regulator | - |
| BAU10_06195 | 1339482..1341638 | - | 2157 | ANP64592 | diguanylate phosphodiesterase | - |
| BAU10_06200 | 1341638..1342567 | - | 930 | ANP64593 | alkylphosphonate ABC transporter | - |
| BAU10_06205 | 1342835..1343560 | - | 726 | ANP64594 | type IV pilus biogenesis/stability protein PilW | - |
| BAU10_06210 | 1343740..1344681 | - | 942 | ANP64595 | homoserine O-succinyltransferase | - |
| BAU10_06215 | 1345022..1345408 | + | 387 | ANP64596 | hypothetical protein | - |
| BAU10_06220 | 1345511..1345867 | + | 357 | ANP64597 | ethidium bromide resistance protein | - |
| BAU10_06225 | 1346288..1347238 | + | 951 | ANP66457 | lipid A biosynthesis lauroyl acyltransferase | - |
| BAU10_06230 | 1347308..1347523 | - | 216 | ANP64598 | hypothetical protein | - |
| BAU10_06235 | 1347623..1348369 | + | 747 | ANP64599 | hypothetical protein | - |
| BAU10_06240 | 1348543..1348764 | + | 222 | ANP64600 | hypothetical protein | - |
| BAU10_06245 | 1348752..1349249 | + | 498 | ANP64601 | hypothetical protein | - |
| BAU10_06250 | 1349328..1351637 | - | 2310 | ANP64602 | hypothetical protein | - |
| BAU10_06255 | 1352885..1353889 | + | 1005 | ANP64603 | hypothetical protein | - |
| BAU10_06260 | 1353902..1354753 | + | 852 | ANP64604 | hypothetical protein | - |
| BAU10_06265 | 1354812..1357433 | + | 2622 | ANP64605 | hypothetical protein | - |
| BAU10_06270 | 1357434..1357916 | - | 483 | ANP64606 | hypothetical protein | - |
| BAU10_06275 | 1357909..1358340 | - | 432 | ANP64607 | hypothetical protein | - |
| BAU10_06280 | 1358925..1359536 | + | 612 | ANP64608 | hypothetical protein | - |
| BAU10_06285 | 1359705..1360034 | + | 330 | ANP64609 | hypothetical protein | - |
| BAU10_06290 | 1360105..1361433 | - | 1329 | ANP64610 | AAA family ATPase | - |
| BAU10_06295 | 1361672..1361962 | + | 291 | ANP64611 | hypothetical protein | - |
| BAU10_06300 | 1362159..1362671 | - | 513 | ANP64612 | hypothetical protein | - |
| BAU10_06305 | 1363223..1366066 | - | 2844 | ANP64613 | hypothetical protein | - |
| BAU10_06310 | 1366059..1367465 | - | 1407 | ANP64614 | hypothetical protein | - |
| BAU10_06315 | 1367465..1368250 | - | 786 | ANP64615 | hypothetical protein | - |
| BAU10_06320 | 1368262..1370076 | - | 1815 | ANP64616 | hypothetical protein | - |
| BAU10_06325 | 1370278..1370502 | + | 225 | ANP64617 | hypothetical protein | - |
| BAU10_06330 | 1370553..1371173 | + | 621 | ANP64618 | hypothetical protein | - |
| BAU10_06335 | 1371315..1372028 | - | 714 | ANP64619 | hypothetical protein | - |
| BAU10_06340 | 1372328..1373119 | - | 792 | ANP64620 | metal-dependent hydrolase | - |
| BAU10_06345 | 1373119..1376385 | - | 3267 | ANP64621 | DEAD/DEAH box helicase | - |
| BAU10_06350 | 1376436..1377293 | - | 858 | ANP64622 | hypothetical protein | - |
| BAU10_06355 | 1377373..1378689 | - | 1317 | ANP64623 | hypothetical protein | - |
| BAU10_06360 | 1378679..1380196 | - | 1518 | ANP64624 | type I restriction-modification system subunit M | - |
| BAU10_06365 | 1380213..1380839 | - | 627 | ANP64625 | hypothetical protein | - |
| BAU10_06370 | 1381065..1382819 | + | 1755 | ANP64626 | hypothetical protein | - |
| BAU10_06375 | 1382803..1383012 | + | 210 | ANP64627 | ATPase | - |
| BAU10_06380 | 1383489..1383887 | + | 399 | ANP64628 | hypothetical protein | - |
| BAU10_06385 | 1383971..1384609 | + | 639 | Protein_1237 | hypothetical protein | - |
| BAU10_06390 | 1384696..1385133 | + | 438 | ANP64629 | AAA family ATPase | - |
| BAU10_06395 | 1385142..1385408 | + | 267 | ANP64630 | hypothetical protein | - |
| BAU10_06400 | 1385463..1385753 | + | 291 | ANP64631 | hypothetical protein | - |
| BAU10_06405 | 1385994..1386728 | + | 735 | ANP64632 | hypothetical protein | - |
| BAU10_06410 | 1386935..1388776 | - | 1842 | ANP64633 | hypothetical protein | - |
| BAU10_06415 | 1389054..1390313 | - | 1260 | ANP64634 | XRE family transcriptional regulator | - |
| BAU10_06420 | 1390313..1390720 | - | 408 | ANP64635 | DNA polymerase V | - |
| BAU10_06425 | 1391142..1391372 | + | 231 | ANP66458 | hypothetical protein | - |
| BAU10_06430 | 1391587..1392966 | - | 1380 | ANP64636 | hypothetical protein | - |
| BAU10_06435 | 1393469..1394311 | - | 843 | ANP64637 | hypothetical protein | - |
| BAU10_06440 | 1394570..1396198 | - | 1629 | ANP64638 | hypothetical protein | - |
| BAU10_06445 | 1396291..1396938 | - | 648 | ANP64639 | hypothetical protein | - |
| BAU10_06450 | 1397761..1398729 | + | 969 | ANP64640 | hypothetical protein | - |
| BAU10_06455 | 1398863..1400410 | - | 1548 | ANP64641 | peptidase M32 | - |
| BAU10_06460 | 1400634..1401269 | + | 636 | ANP66459 | threonine transporter RhtB | - |
| BAU10_06465 | 1401269..1401835 | + | 567 | ANP64642 | threonine transporter RhtB | - |
| BAU10_06470 | 1401954..1403330 | - | 1377 | ANP64643 | sodium:alanine symporter | - |
| BAU10_06475 | 1404270..1405004 | + | 735 | ANP64644 | chromosome partitioning protein ParA | - |
| BAU10_06480 | 1405094..1405477 | - | 384 | ANP64645 | hypothetical protein | - |
| BAU10_06485 | 1405577..1405942 | - | 366 | ANP64646 | glyoxalase | - |
| BAU10_06490 | 1406089..1408038 | - | 1950 | ANP64647 | hypothetical protein | - |
| BAU10_06495 | 1408233..1409240 | - | 1008 | ANP64648 | glycine betaine ABC transporter substrate-binding protein | - |
| BAU10_06500 | 1409233..1410318 | - | 1086 | ANP64649 | glycine/betaine ABC transporter permease | - |
| BAU10_06505 | 1410323..1411510 | - | 1188 | ANP64650 | glycine betaine/L-proline ABC transporter ATP-binding protein | - |
| BAU10_06510 | 1411949..1413829 | + | 1881 | ANP64651 | cyclic nucleotide-binding protein | - |
| BAU10_06515 | 1413826..1414575 | + | 750 | ANP64652 | DNA polymerase III subunit epsilon | - |
| BAU10_06520 | 1414769..1416427 | + | 1659 | ANP64653 | BCCT transporter | - |
| BAU10_06525 | 1416742..1417275 | + | 534 | ANP64654 | diaminobutyrate acetyltransferase | - |
| BAU10_06530 | 1417310..1418575 | + | 1266 | ANP64655 | diaminobutyrate--2-oxoglutarate transaminase | - |
| BAU10_06535 | 1418590..1418976 | + | 387 | ANP64656 | L-ectoine synthase | - |
| BAU10_06540 | 1419063..1420484 | + | 1422 | ANP64657 | aspartate kinase | - |
| BAU10_06545 | 1420673..1422301 | - | 1629 | ANP64658 | arylsulfatase | - |
| BAU10_06550 | 1422628..1424958 | + | 2331 | ANP64659 | deca-heme c-type cytochrome | - |
| BAU10_06555 | 1425004..1425792 | - | 789 | ANP64660 | sulfatase | - |
| BAU10_06560 | 1425971..1427296 | + | 1326 | ANP64661 | anaerobic sulfatase maturase | - |
| BAU10_06565 | 1427350..1428447 | + | 1098 | ANP64662 | hypothetical protein | - |
| BAU10_06570 | 1428610..1429608 | + | 999 | ANP64663 | haloacid dehalogenase | - |
| BAU10_06575 | 1429716..1431233 | - | 1518 | ANP64664 | arylsulfatase | - |
| BAU10_06580 | 1431466..1432917 | + | 1452 | ANP64665 | arylsulfatase | - |
| BAU10_06585 | 1433070..1445210 | + | 12141 | ANP64666 | RTX toxin | - |
| BAU10_06590 | 1445207..1447378 | + | 2172 | ANP64667 | peptidase C39 | - |
| BAU10_06595 | 1447375..1448688 | + | 1314 | ANP64668 | hemolysin D | - |
| BAU10_06600 | 1448704..1450017 | + | 1314 | ANP64669 | agglutination protein | - |
| BAU10_06605 | 1450010..1450630 | + | 621 | ANP64670 | hypothetical protein | - |
| BAU10_06610 | 1450620..1451462 | + | 843 | ANP64671 | phosphate ABC transporter substrate-binding protein | - |
| BAU10_06615 | 1451641..1452414 | + | 774 | ANP64672 | DeoR family transcriptional regulator | - |
| BAU10_06620 | 1452426..1453466 | + | 1041 | ANP64673 | oxidoreductase | - |
| BAU10_06625 | 1453463..1453933 | + | 471 | ANP64674 | hypothetical protein | - |
| BAU10_06630 | 1454118..1455695 | + | 1578 | ANP64675 | diguanylate cyclase | - |
| BAU10_06635 | 1455747..1456304 | - | 558 | ANP64676 | hypothetical protein | - |
| BAU10_06640 | 1456784..1457146 | + | 363 | ANP64677 | hypothetical protein | - |
| BAU10_06645 | 1457182..1457646 | + | 465 | ANP64678 | acetyltransferase | - |
| BAU10_06650 | 1457895..1459313 | + | 1419 | ANP64679 | dihydroorotate dehydrogenase | - |
| BAU10_06655 | 1459317..1460654 | + | 1338 | ANP64680 | histidine kinase | - |
| BAU10_06660 | 1460641..1461114 | + | 474 | ANP64681 | hypothetical protein | - |
| BAU10_06665 | 1461222..1462925 | - | 1704 | ANP64682 | histidine kinase | - |
| BAU10_06670 | 1463413..1464630 | + | 1218 | ANP64683 | HD family phosphohydrolase | - |
| BAU10_06675 | 1464833..1465456 | + | 624 | ANP64684 | flagellar biosynthesis protein FlgM | - |
| BAU10_06680 | 1465668..1467212 | + | 1545 | ANP64685 | YjjI family glycine radical enzyme | - |
| BAU10_06685 | 1467235..1468116 | + | 882 | ANP64686 | glycine radical enzyme activase | - |
| BAU10_06690 | 1468136..1469074 | - | 939 | ANP64687 | LysR family transcriptional regulator | - |
| BAU10_06695 | 1469486..1470256 | + | 771 | ANP64688 | hypothetical protein | - |
| BAU10_06700 | 1470417..1471019 | + | 603 | ANP64689 | SM-20 protein | - |
| BAU10_06705 | 1471074..1472339 | - | 1266 | ANP64690 | hydrolase | - |
| BAU10_06710 | 1472969..1474162 | + | 1194 | ANP64691 | alcohol dehydrogenase | - |
| BAU10_06715 | 1474324..1475529 | + | 1206 | ANP64692 | tyrosine transporter TyrP | - |
| BAU10_06720 | 1475804..1476928 | + | 1125 | ANP64693 | c-di-GMP phosphodiesterase | - |
| BAU10_06725 | 1477016..1477234 | + | 219 | ANP64694 | hypothetical protein | - |
| BAU10_06730 | 1477559..1478323 | + | 765 | ANP64695 | iron-hydroxamate transporter ATP-binding subunit | - |
| BAU10_06735 | 1478335..1479219 | + | 885 | ANP64696 | ABC transporter substrate-binding protein | - |
| BAU10_06740 | 1479216..1481192 | + | 1977 | ANP64697 | Fe3+-hydroxamate ABC transporter permease FhuB | - |
| BAU10_06745 | 1481393..1483615 | + | 2223 | ANP64698 | ligand-gated channel protein | - |
| BAU10_06750 | 1483658..1484206 | - | 549 | ANP64699 | hypothetical protein | - |
| BAU10_06755 | 1484458..1486233 | - | 1776 | ANP64700 | multidrug ABC transporter permease/ATP-binding protein | - |
| BAU10_06760 | 1486237..1487976 | - | 1740 | ANP64701 | multidrug ABC transporter permease/ATP-binding protein | - |
| BAU10_06765 | 1488248..1488739 | - | 492 | ANP64702 | V10 pilin | - |
| BAU10_06770 | 1488944..1490287 | - | 1344 | ANP64703 | sodium:glutamate symporter | - |
| BAU10_06775 | 1490714..1491592 | - | 879 | ANP64704 | LysR family transcriptional regulator | - |
| BAU10_06780 | 1491696..1491983 | + | 288 | ANP64705 | antibiotic biosynthesis monooxygenase | - |
| BAU10_06785 | 1491995..1492972 | + | 978 | ANP64706 | oxidoreductase | - |
| BAU10_06790 | 1493057..1493743 | - | 687 | ANP64707 | 3-methyladenine DNA glycosylase | - |
| BAU10_06795 | 1493830..1494024 | + | 195 | ANP64708 | hypothetical protein | - |
| BAU10_06800 | 1494110..1496920 | - | 2811 | ANP64709 | molecular chaperone DnaK | - |
| BAU10_06805 | 1496976..1498940 | - | 1965 | ANP64710 | molecular chaperone DnaK | - |
| BAU10_06810 | 1498940..1499572 | - | 633 | ANP64711 | hypothetical protein | - |
| BAU10_06815 | 1499864..1501405 | + | 1542 | ANP64712 | diguanylate cyclase | - |
| BAU10_06820 | 1501489..1502019 | + | 531 | ANP64713 | exonuclease | - |
| BAU10_06825 | 1502261..1502701 | - | 441 | ANP64714 | hypothetical protein | - |
| BAU10_06830 | 1502892..1504832 | - | 1941 | ANP64715 | hypothetical protein | - |
| BAU10_06835 | 1504842..1505192 | - | 351 | ANP64716 | hypothetical protein | - |
| BAU10_06840 | 1505428..1506732 | + | 1305 | ANP66460 | MltA-interacting MipA family protein | - |
| BAU10_06845 | 1506810..1507184 | + | 375 | ANP66461 | hypothetical protein | - |
| BAU10_06850 | 1507279..1507863 | - | 585 | ANP64717 | FMN-dependent NADH-azoreductase | - |
| BAU10_06855 | 1508199..1512035 | + | 3837 | ANP66462 | ATP-dependent RNA helicase HrpA | - |
| BAU10_06860 | 1512218..1512697 | + | 480 | ANP66463 | hypothetical protein | - |
| BAU10_06865 | 1512897..1513412 | + | 516 | ANP64718 | hypothetical protein | - |
| BAU10_06870 | 1513775..1515355 | + | 1581 | ANP64719 | BCCT transporter | - |
| BAU10_06875 | 1515482..1515871 | + | 390 | ANP64720 | shikimate kinase | - |
| BAU10_06880 | 1515891..1516529 | - | 639 | ANP64721 | sugar transferase | - |
| BAU10_06885 | 1516540..1517730 | - | 1191 | ANP64722 | glycosyl transferase | - |
| BAU10_06890 | 1517723..1518841 | - | 1119 | ANP64723 | glycosyl transferase | - |
| BAU10_06895 | 1519063..1520496 | - | 1434 | ANP64724 | chain-length determining protein | - |
| BAU10_06900 | 1520493..1521617 | - | 1125 | ANP64725 | glycosyl transferase | - |
| BAU10_06905 | 1521733..1523097 | - | 1365 | ANP64726 | hypothetical protein | - |
| BAU10_06910 | 1523094..1524524 | - | 1431 | ANP64727 | lipopolysaccharide biosynthesis protein | - |
| BAU10_06915 | 1524517..1525689 | - | 1173 | ANP64728 | glycosyltransferase | - |
| BAU10_06920 | 1525701..1526885 | - | 1185 | ANP64729 | glycosyl transferase | - |
| BAU10_06925 | 1527030..1528100 | - | 1071 | ANP64730 | glycosyl transferase family 1 | - |
| BAU10_06930 | 1528075..1529601 | - | 1527 | ANP64731 | sigma-54-dependent Fis family transcriptional regulator | - |
| BAU10_06935 | 1529910..1530290 | - | 381 | ANP64732 | phosphorelay protein LuxU | - |
| BAU10_06940 | 1530302..1530988 | - | 687 | ANP64733 | chromosome partitioning protein ParA | - |
| BAU10_06945 | 1530988..1533090 | - | 2103 | ANP64734 | sugar ABC transporter substrate-binding protein | - |
| BAU10_06950 | 1533087..1533953 | - | 867 | ANP64735 | hypothetical protein | - |
| BAU10_06955 | 1533980..1534300 | - | 321 | ANP64736 | anti-anti-sigma factor | - |
| BAU10_06960 | 1534724..1535569 | + | 846 | ANP64737 | DNA ligase | - |
| BAU10_06965 | 1535585..1536583 | - | 999 | ANP64738 | hypothetical protein | - |
| BAU10_06970 | 1536718..1538088 | - | 1371 | ANP64739 | MATE family efflux transporter | - |
| BAU10_06975 | 1538481..1539092 | + | 612 | ANP64740 | riboflavin synthase subunit alpha | - |
| BAU10_06980 | 1539168..1539362 | - | 195 | ANP64741 | molybdopterin-guanine dinucleotide biosynthesis protein A | - |
| BAU10_06985 | 1539616..1540482 | - | 867 | ANP64742 | hypothetical protein | - |
| BAU10_06990 | 1540707..1541681 | + | 975 | ANP66464 | chemotaxis protein CheC | - |
| BAU10_06995 | 1541681..1542643 | + | 963 | ANP64743 | diguanylate cyclase | - |
| BAU10_07000 | 1542834..1543178 | + | 345 | ANP64744 | topoisomerase II | - |
| BAU10_07005 | 1543224..1543718 | - | 495 | ANP66465 | hydrolase | - |
| BAU10_07010 | 1543853..1545871 | + | 2019 | ANP64745 | chemotaxis protein | - |
| BAU10_07015 | 1546142..1546555 | + | 414 | ANP66466 | DNA mismatch repair protein MutT | - |
| BAU10_07020 | 1546766..1547146 | + | 381 | ANP64746 | hypothetical protein | - |
| BAU10_07025 | 1547195..1547413 | + | 219 | ANP64747 | hypothetical protein | - |
| BAU10_07030 | 1547517..1548332 | - | 816 | ANP66467 | sterol desaturase | - |
| BAU10_07035 | 1548473..1549222 | + | 750 | ANP64748 | DTW domain-containing protein | - |
| BAU10_07040 | 1549398..1550306 | + | 909 | ANP64749 | N-acetylglucosamine kinase | - |
| BAU10_07045 | 1550533..1550799 | - | 267 | ANP64750 | DUF2960 domain-containing protein | - |
| BAU10_07050 | 1550874..1552652 | - | 1779 | ANP64751 | bifunctional molybdopterin-guanine dinucleotide biosynthesis protein MobB/molybdopterin molybdotransferase MoeA | - |
| BAU10_07055 | 1552649..1553236 | - | 588 | ANP64752 | molybdenum cofactor guanylyltransferase MobA | - |
| BAU10_07060 | 1553214..1553933 | - | 720 | ANP64753 | ABC transporter ATP-binding protein | - |
| BAU10_07065 | 1553984..1554682 | - | 699 | ANP64754 | ABC transporter permease | - |
| BAU10_07070 | 1554689..1555507 | - | 819 | ANP64755 | tungsten ABC transporter substrate-binding protein | - |
| BAU10_07075 | 1555842..1557293 | + | 1452 | ANP64756 | sigma-54-dependent Fis family transcriptional regulator | - |
| BAU10_07080 | 1557283..1559322 | + | 2040 | ANP64757 | two-component sensor histidine kinase | - |
| BAU10_07085 | 1559539..1561131 | + | 1593 | ANP64758 | ABC-F family ATPase | - |
| BAU10_07090 | 1561372..1562196 | - | 825 | ANP64759 | formate dehydrogenase family accessory protein FdhD | - |
| BAU10_07095 | 1562520..1562984 | + | 465 | ANP64760 | hypothetical protein | - |
| BAU10_07100 | 1562974..1563603 | + | 630 | ANP64761 | hypothetical protein | - |
| BAU10_07105 | 1563912..1565573 | + | 1662 | ANP64762 | (Fe-S)-binding protein | - |
| BAU10_07110 | 1565583..1566209 | + | 627 | ANP64763 | hypothetical protein | - |
| BAU10_07115 | 1566283..1566483 | + | 201 | ANP64764 | transcriptional initiation protein Tat | - |
| BAU10_07120 | 1566494..1569349 | + | 2856 | ANP64765 | formate dehydrogenase | - |
| BAU10_07125 | 1569361..1569969 | + | 609 | ANP64766 | formate dehydrogenase | - |
| BAU10_07130 | 1569984..1571036 | + | 1053 | ANP64767 | formate dehydrogenase subunit gamma | - |
| BAU10_07135 | 1571020..1571436 | + | 417 | ANP64768 | hypothetical protein | - |
| BAU10_07140 | 1571860..1576239 | + | 4380 | ANP64769 | type IV secretion protein Rhs | - |
| BAU10_07145 | 1576256..1577137 | + | 882 | ANP64770 | hypothetical protein | - |
| BAU10_07150 | 1577460..1578680 | + | 1221 | ANP64771 | ammonium transporter | - |
| BAU10_07155 | 1579005..1579736 | + | 732 | ANP64772 | NAD-dependent protein deacylase | - |
| BAU10_07160 | 1579825..1580862 | - | 1038 | ANP64773 | spermidine/putrescine ABC transporter substrate-binding protein PotD | - |
| BAU10_07165 | 1581112..1582143 | - | 1032 | ANP64774 | spermidine/putrescine ABC transporter substrate-binding protein PotD | - |
| BAU10_07170 | 1582246..1583016 | - | 771 | ANP64775 | spermidine/putrescine ABC transporter permease PotC | - |
| BAU10_07175 | 1583016..1583876 | - | 861 | ANP64776 | spermidine/putrescine ABC transporter permease PotB | - |
| BAU10_07180 | 1583857..1584972 | - | 1116 | ANP64777 | putrescine/spermidine ABC transporter ATP-binding protein | - |
| BAU10_07185 | 1585373..1586170 | + | 798 | ANP64778 | hypothetical protein | - |
| BAU10_07190 | 1586163..1586999 | + | 837 | ANP64779 | glucosaminidase | - |
| BAU10_07195 | 1587013..1587675 | + | 663 | ANP64780 | hypothetical protein | - |
| BAU10_07200 | 1587874..1588767 | + | 894 | ANP64781 | tRNA 2-thiocytidine(32) synthetase TtcA | - |
| BAU10_07205 | 1588896..1589843 | - | 948 | ANP64782 | universal stress protein UspE | - |
| BAU10_07210 | 1590024..1590770 | - | 747 | ANP64783 | transcriptional regulator | - |
| BAU10_07215 | 1590861..1591541 | - | 681 | ANP64784 | cytochrome biogenesis protein | - |
| BAU10_07220 | 1591549..1591713 | - | 165 | ANP64785 | cytochrome oxidase maturation protein, cbb3-type | - |
| BAU10_07225 | 1591729..1594092 | - | 2364 | ANP64786 | ATPase P | - |
| BAU10_07230 | 1594097..1594573 | - | 477 | ANP64787 | hypothetical protein | - |
| BAU10_07235 | 1594679..1595653 | - | 975 | ANP64788 | cytochrome c oxidase, cbb3-type subunit III | - |
| BAU10_07240 | 1595653..1595826 | - | 174 | ANP64789 | cytochrome-c oxidase | - |
| BAU10_07245 | 1595836..1596456 | - | 621 | ANP64790 | cytochrome c oxidase, cbb3-type subunit II | - |
| BAU10_07250 | 1596469..1597896 | - | 1428 | ANP64791 | cytochrome c oxidase, cbb3-type subunit I | - |
| BAU10_07255 | 1598154..1598510 | + | 357 | ANP64792 | hypothetical protein | - |
| BAU10_07260 | 1598624..1599805 | + | 1182 | ANP64793 | histidine kinase | - |
| BAU10_07265 | 1599795..1601516 | + | 1722 | ANP64794 | hybrid sensor histidine kinase/response regulator | - |
| BAU10_07270 | 1602137..1602460 | - | 324 | ANP64795 | hypothetical protein | - |
| BAU10_07275 | 1602576..1602887 | - | 312 | ANP64796 | hypothetical protein | - |
| BAU10_07280 | 1602903..1603352 | - | 450 | ANP64797 | macrodomain Ter protein | - |
| BAU10_07285 | 1603563..1605203 | + | 1641 | ANP64798 | ATP-dependent protease | - |
| BAU10_07290 | 1605274..1605792 | + | 519 | ANP64799 | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabA | - |
| BAU10_07295 | 1605933..1606106 | - | 174 | ANP64800 | ribosome modulation factor | - |
| BAU10_07300 | 1606288..1608021 | - | 1734 | ANP64801 | hypothetical protein | - |
| BAU10_07305 | 1608029..1609948 | - | 1920 | ANP64802 | ABC transporter ATP-binding protein | - |
| BAU10_07310 | 1609926..1610126 | - | 201 | ANP64803 | thiol-disulfide isomerase | - |
| BAU10_07315 | 1610190..1612310 | - | 2121 | ANP64804 | 23S rRNA (guanine(2445)-N(2))/(guanine(2069)-N(7))- methyltransferase | - |
| BAU10_07320 | 1612797..1613339 | - | 543 | ANP64805 | cell division protein ZapC | - |
| BAU10_07325 | 1613508..1614518 | - | 1011 | ANP64806 | dihydroorotate dehydrogenase (quinone) | - |
| BAU10_07330 | 1614830..1619671 | - | 4842 | ANP64807 | NAD-glutamate dehydrogenase | - |
| BAU10_07335 | 1620157..1622763 | - | 2607 | ANP64808 | aminopeptidase N | - |
| BAU10_07340 | 1622990..1623655 | - | 666 | ANP64809 | hypothetical protein | - |
| BAU10_07345 | 1623850..1625856 | - | 2007 | ANP64810 | C-terminal processing peptidase | - |
| BAU10_07350 | 1625874..1626500 | - | 627 | ANP64811 | RNA chaperone ProQ | - |
| BAU10_07355 | 1626598..1627071 | - | 474 | ANP64812 | Free methionine-(R)-sulfoxide reductase | - |
| BAU10_07360 | 1627164..1628960 | + | 1797 | ANP64813 | ABC transporter ATP-binding protein | - |
| BAU10_07365 | 1628985..1630235 | + | 1251 | ANP64814 | paraquat-inducible protein A | - |
| BAU10_07370 | 1630228..1632876 | + | 2649 | ANP64815 | paraquat-inducible protein B | - |
| BAU10_07375 | 1633027..1634463 | + | 1437 | ANP64816 | 16S rRNA (cytosine(1407)-C(5))-methyltransferase RsmF | - |
| BAU10_07380 | 1634746..1635579 | - | 834 | ANP64817 | GntR family transcriptional regulator | - |
| BAU10_07385 | 1635772..1637124 | + | 1353 | ANP64818 | hypothetical protein | - |
| BAU10_07390 | 1637158..1637655 | - | 498 | ANP64819 | DUF4442 domain-containing protein | - |
| BAU10_07395 | 1638028..1639086 | + | 1059 | ANP64820 | 3-deoxy-7-phosphoheptulonate synthase | - |
| BAU10_07400 | 1639108..1639590 | + | 483 | ANP64821 | YajQ family cyclic di-GMP-binding protein | - |
| BAU10_07405 | 1639693..1640613 | - | 921 | ANP64822 | ferrochelatase | - |
| BAU10_07410 | 1641074..1642102 | + | 1029 | ANP64823 | amino acid ABC transporter substrate-binding protein | - |
| BAU10_07415 | 1642320..1643525 | + | 1206 | ANP64824 | amino acid ABC transporter permease | - |
| BAU10_07420 | 1643527..1644624 | + | 1098 | ANP64825 | amino acid ABC transporter permease | - |
| BAU10_07425 | 1644648..1645397 | + | 750 | ANP64826 | ABC transporter ATP-binding protein | - |
| BAU10_07430 | 1645646..1646314 | + | 669 | ANP64827 | BAX inhibitor protein | - |
| BAU10_07435 | 1646391..1646765 | - | 375 | ANP64828 | chromosome partitioning protein ParB | - |
| BAU10_07440 | 1646957..1647295 | - | 339 | ANP64829 | hypothetical protein | - |
| BAU10_07445 | 1647674..1648003 | + | 330 | ANP64830 | sulfur relay protein TusE | - |
| BAU10_07450 | 1648006..1648278 | - | 273 | ANP64831 | acylphosphatase | - |
| BAU10_07455 | 1648369..1649772 | + | 1404 | ANP64832 | chemotaxis protein | - |
| BAU10_07460 | 1649933..1651126 | + | 1194 | ANP64833 | 23S rRNA methyltransferase | - |
| BAU10_07465 | 1651179..1658228 | - | 7050 | Protein_1453 | calcium-binding protein | - |
| BAU10_07470 | 1658265..1662944 | - | 4680 | Protein_1454 | calcium-binding protein | - |
| BAU10_07475 | 1663361..1664674 | + | 1314 | ANP64834 | agglutination protein | - |
| BAU10_07480 | 1664677..1665291 | + | 615 | ANP64835 | hypothetical protein | - |
| BAU10_07485 | 1665379..1690464 | - | 25086 | ANP64836 | hypothetical protein | - |
| BAU10_07490 | 1690865..1692175 | + | 1311 | ANP64837 | agglutination protein | - |
| BAU10_07495 | 1692178..1692792 | + | 615 | ANP64838 | hypothetical protein | - |
| BAU10_07500 | 1692968..1693321 | + | 354 | ANP64839 | hypothetical protein | - |
| BAU10_07505 | 1693508..1695478 | + | 1971 | ANP64840 | diguanylate cyclase | - |
| BAU10_07510 | 1695566..1696264 | + | 699 | ANP64841 | transcriptional regulator | - |
| BAU10_07515 | 1696371..1697063 | + | 693 | ANP64842 | SDR family oxidoreductase | - |
| BAU10_07520 | 1697262..1698062 | - | 801 | ANP64843 | 2,5-didehydrogluconate reductase B | - |
| BAU10_07525 | 1698175..1699080 | + | 906 | ANP64844 | LysR family transcriptional regulator | - |
| BAU10_07530 | 1699213..1700118 | + | 906 | ANP64845 | LLM class oxidoreductase | - |
| BAU10_07535 | 1700234..1701118 | - | 885 | ANP64846 | LysR family transcriptional regulator | - |
| BAU10_07540 | 1701278..1702459 | + | 1182 | ANP64847 | MFS sugar transporter | - |
| BAU10_07545 | 1702603..1703124 | + | 522 | ANP66468 | TetR family transcriptional regulator | - |
| BAU10_07550 | 1703121..1704170 | + | 1050 | ANP64848 | alcohol dehydrogenase | - |
| BAU10_07555 | 1704374..1705285 | - | 912 | ANP64849 | AraC family transcriptional regulator | - |
| BAU10_07560 | 1705406..1705924 | + | 519 | ANP64850 | isochorismatase | - |
| BAU10_07565 | 1706045..1706806 | + | 762 | ANP64851 | nickel transporter | - |
| BAU10_07570 | 1706905..1707999 | - | 1095 | ANP64852 | hypothetical protein | - |
| BAU10_07575 | 1708325..1709317 | + | 993 | ANP64853 | dehydrogenase | - |
| BAU10_07580 | 1709465..1710367 | - | 903 | ANP64854 | homocysteine methyltransferase | - |
| BAU10_07585 | 1710643..1711368 | - | 726 | ANP66469 | peptide ABC transporter ATP-binding protein | - |
| BAU10_07590 | 1711392..1712339 | - | 948 | ANP64855 | ABC transporter permease | - |
| BAU10_07595 | 1712475..1713263 | - | 789 | ANP64856 | amino acid ABC transporter substrate-binding protein | - |
| BAU10_07600 | 1713725..1714069 | + | 345 | ANP64857 | acyl-CoA synthetase | - |
| BAU10_07605 | 1714674..1715648 | - | 975 | ANP64858 | outer membrane protein C precursor | - |
| BAU10_07610 | 1715789..1716712 | - | 924 | ANP64859 | LysR family transcriptional regulator | - |
| BAU10_07615 | 1717091..1718260 | + | 1170 | ANP64860 | efflux transporter periplasmic adaptor subunit | - |
| BAU10_07620 | 1718273..1721413 | + | 3141 | ANP64861 | multidrug efflux RND transporter permease subunit | - |
| BAU10_07625 | 1721559..1722488 | + | 930 | ANP64862 | LysR family transcriptional regulator | - |
| BAU10_07630 | 1722683..1723294 | + | 612 | ANP64863 | hypothetical protein | - |
| BAU10_07635 | 1723717..1724616 | - | 900 | ANP64864 | LysR family transcriptional regulator | - |
| BAU10_07640 | 1724729..1725313 | + | 585 | ANP64865 | amino acid transporter | - |
| BAU10_07645 | 1725622..1725963 | + | 342 | ANP64866 | transcriptional regulator | - |
| BAU10_07650 | 1726031..1726687 | + | 657 | ANP64867 | 7-cyano-7-deazaguanine synthase QueC | - |
| BAU10_07655 | 1726997..1727647 | - | 651 | ANP66470 | 7-carboxy-7-deazaguanine synthase QueE | - |
| BAU10_07660 | 1727864..1728673 | + | 810 | ANP64868 | HAD family hydrolase | - |
| BAU10_07665 | 1728834..1730279 | - | 1446 | ANP64869 | DNA mismatch repair protein MutT | - |
| BAU10_07670 | 1730665..1731996 | - | 1332 | ANP64870 | trypsin | - |
| BAU10_07675 | 1732188..1732667 | - | 480 | ANP64871 | histone acetyltransferase | - |
| BAU10_07680 | 1732857..1734734 | - | 1878 | ANP64872 | propionyl-CoA synthetase | - |
| BAU10_07685 | 1734812..1736008 | - | 1197 | ANP64873 | putative AcnD-accessory protein PrpF | - |
| BAU10_07690 | 1736018..1738609 | - | 2592 | ANP64874 | Fe/S-dependent 2-methylisocitrate dehydratase AcnD | - |
| BAU10_07695 | 1738766..1739908 | - | 1143 | ANP64875 | 2-methylcitrate synthase | - |
| BAU10_07700 | 1740041..1740937 | - | 897 | ANP64876 | methylisocitrate lyase | - |
| BAU10_07705 | 1740950..1741663 | - | 714 | ANP64877 | GntR family transcriptional regulator | - |
| BAU10_07710 | 1741927..1742325 | - | 399 | ANP64878 | aldehyde-activating protein | - |
| BAU10_07715 | 1742591..1743562 | + | 972 | ANP64879 | transporter | - |
| BAU10_07720 | 1743801..1744295 | + | 495 | ANP64880 | hypothetical protein | - |
| BAU10_07725 | 1744315..1745841 | + | 1527 | ANP64881 | tripartite tricarboxylate transporter TctA | - |
| BAU10_07730 | 1746121..1746636 | + | 516 | Protein_1506 | adenylate kinase | - |
| BAU10_07735 | 1746718..1747488 | + | 771 | ANP64882 | permease | - |
| BAU10_07740 | 1747782..1748753 | - | 972 | ANP64883 | C4-dicarboxylate ABC transporter substrate-binding protein | - |
| BAU10_07745 | 1749013..1749378 | - | 366 | ANP64884 | hypothetical protein | - |
| BAU10_07750 | 1749732..1750736 | - | 1005 | ANP64885 | translocator protein PopD | - |
| BAU10_07755 | 1750747..1751946 | - | 1200 | ANP64886 | translocator protein PopB | - |
| BAU10_07760 | 1751954..1752439 | - | 486 | ANP64887 | CesD/SycD/LcrH family type III secretion system chaperone | - |
| BAU10_07765 | 1752452..1754275 | - | 1824 | ANP64888 | Type III secretion cytoplasmic LcrG inhibitor | - |
| BAU10_07770 | 1754286..1754570 | - | 285 | ANP64889 | type III secretion protein LcrG | - |
| BAU10_07775 | 1754631..1755044 | - | 414 | ANP64890 | type III secretion system regulator LcrR | - |
| BAU10_07780 | 1755044..1757161 | - | 2118 | ANP64891 | EscV/YscV/HrcV family type III secretion system export apparatus protein | - |
| BAU10_07785 | 1757154..1757495 | - | 342 | ANP64892 | translocation protein Y | - |
| BAU10_07790 | 1757501..1757878 | - | 378 | ANP64893 | type III secretion protein, YscX family | - |
| BAU10_07795 | 1757875..1758249 | - | 375 | ANP64894 | type III secretion chaperone SycN | - |
| BAU10_07800 | 1758236..1758520 | - | 285 | ANP64895 | SepL/TyeA/HrpJ family type III secretion system gatekeeper | - |
| BAU10_07805 | 1758501..1759394 | - | 894 | ANP64896 | hypothetical protein | - |
| BAU10_07810 | 1759580..1760902 | + | 1323 | ANP64897 | EscN/YscN/HrcN family type III secretion system ATPase | - |
| BAU10_07815 | 1760899..1761360 | + | 462 | ANP64898 | type III secretion protein | - |
| BAU10_07820 | 1761370..1762575 | + | 1206 | ANP64899 | type III secretion system needle length determinant | - |
| BAU10_07825 | 1762572..1763540 | + | 969 | ANP64900 | type III secretion system protein | - |
| BAU10_07830 | 1763537..1764187 | + | 651 | ANP64901 | EscR/YscR/HrcR family type III secretion system export apparatus protein | - |
| BAU10_07835 | 1764201..1764467 | + | 267 | ANP64902 | EscS/YscS/HrcS family type III secretion system export apparatus protein | - |
| BAU10_07840 | 1764464..1765249 | + | 786 | ANP64903 | EscT/YscT/HrcT family type III secretion system export apparatus protein | - |
| BAU10_07845 | 1765265..1766314 | + | 1050 | ANP64904 | EscU/YscU/HrcU family type III secretion system export apparatus switch protein | - |
| BAU10_07850 | 1766417..1767127 | + | 711 | ANP64905 | AraC family transcriptional regulator | - |
| BAU10_07855 | 1767240..1768136 | + | 897 | ANP64906 | hypothetical protein | - |
| BAU10_07860 | 1768226..1769704 | - | 1479 | ANP64907 | cation transporter | - |
| BAU10_07865 | 1769714..1770172 | - | 459 | ANP64908 | type III secretion chaperone CesT | - |
| BAU10_07870 | 1770412..1770858 | - | 447 | ANP64909 | type III secretion chaperone CesT | - |
| BAU10_07875 | 1771114..1772277 | - | 1164 | ANP64910 | effector protein | - |
| BAU10_07880 | 1772281..1772712 | - | 432 | ANP64911 | type III chaperone | - |
| BAU10_07885 | 1772898..1773536 | - | 639 | ANP64912 | type III secretion system protein | - |
| BAU10_07890 | 1773515..1774180 | - | 666 | ANP64913 | type III secretion protein | - |
| BAU10_07895 | 1774173..1774916 | - | 744 | ANP64914 | EscJ/YscJ/HrcJ family type III secretion inner membrane ring protein | - |
| BAU10_07900 | 1774919..1775263 | - | 345 | ANP64915 | EscI/YscI/HrpB family type III secretion system inner rod protein | - |
| BAU10_07905 | 1775272..1775907 | - | 636 | ANP64916 | type III secretion effector, YopR family | - |
| BAU10_07910 | 1775904..1776263 | - | 360 | ANP64917 | type III secretion protein, YscG family | - |
| BAU10_07915 | 1776267..1776515 | - | 249 | ANP64918 | EscF/YscF/HrpA family type III secretion system needle major subunit | - |
| BAU10_07920 | 1776534..1776737 | - | 204 | ANP64919 | EscE/YscE/SsaE family type III secretion system needle protein co-chaperone | - |
| BAU10_07925 | 1776724..1778025 | - | 1302 | ANP64920 | EscD/YscD/HrpQ family type III secretion system inner membrane ring protein | - |
| BAU10_07930 | 1778031..1779905 | - | 1875 | ANP64921 | EscC/YscC/HrcC family type III secretion system outer membrane ring protein | - |
| BAU10_07935 | 1779902..1780330 | - | 429 | ANP64922 | type III secretion system chaperone, YscB family | - |
| BAU10_07940 | 1780343..1781323 | - | 981 | ANP64923 | Type III secretion negative regulator (LscZ) | - |
| BAU10_07945 | 1781472..1782332 | - | 861 | ANP64924 | transcriptional regulator | - |
| BAU10_07950 | 1782942..1783355 | - | 414 | ANP64925 | type III secretion system chaperone YscW | - |
| BAU10_07955 | 1783744..1784190 | + | 447 | ANP64926 | exoenzyme S synthesis protein C | - |
| BAU10_07960 | 1784199..1784504 | + | 306 | ANP64927 | Type III secretion regulator ExsE | - |
| BAU10_07965 | 1784631..1786151 | - | 1521 | ANP64928 | aldehyde dehydrogenase | - |
| BAU10_07970 | 1786391..1788157 | + | 1767 | ANP64929 | sigma-54-dependent Fis family transcriptional regulator | - |
| BAU10_07975 | 1788935..1789138 | + | 204 | ANP64930 | hypothetical protein | - |
| BAU10_07980 | 1789149..1789676 | - | 528 | ANP64931 | hypothetical protein | - |
| BAU10_07985 | 1789673..1790527 | - | 855 | ANP64932 | hypothetical protein | - |
| BAU10_07990 | 1790725..1792173 | - | 1449 | ANP64933 | phosphogluconate dehydrogenase (NADP(+)-dependent, decarboxylating) | - |
| BAU10_07995 | 1792212..1792928 | - | 717 | ANP64934 | 6-phosphogluconolactonase | - |
| BAU10_08000 | 1792925..1794427 | - | 1503 | ANP64935 | glucose-6-phosphate dehydrogenase | - |
| BAU10_08005 | 1794965..1795930 | + | 966 | ANP64936 | AraC family transcriptional regulator | - |
| BAU10_08010 | 1796031..1798166 | + | 2136 | ANP64937 | ligand-gated channel protein | - |
| BAU10_08015 | 1798258..1798944 | - | 687 | ANP64938 | two-component system response regulator | - |
| BAU10_08020 | 1798941..1800560 | - | 1620 | ANP64939 | histidine kinase | - |
| BAU10_08025 | 1801037..1801264 | + | 228 | ANP64940 | hypothetical protein | - |
| BAU10_08030 | 1801395..1802810 | + | 1416 | ANP64941 | hypothetical protein | - |
| BAU10_08035 | 1802814..1803794 | + | 981 | ANP64942 | hypothetical protein | - |
| BAU10_08040 | 1803817..1804983 | + | 1167 | ANP64943 | multidrug ABC transporter permease | - |
| BAU10_08045 | 1804980..1806131 | + | 1152 | ANP64944 | ABC transporter | - |
| BAU10_08050 | 1806418..1807326 | - | 909 | ANP64945 | LysR family transcriptional regulator | - |
| BAU10_08055 | 1807469..1807897 | - | 429 | ANP64946 | hypothetical protein | - |
| BAU10_08060 | 1807915..1809084 | - | 1170 | ANP64947 | phospholipase | - |
| BAU10_08065 | 1809295..1810152 | - | 858 | ANP64948 | pyridoxal kinase | - |
| BAU10_08070 | 1810278..1811873 | - | 1596 | ANP64949 | aspartate 4-decarboxylase | - |
| BAU10_08075 | 1811969..1813684 | - | 1716 | ANP64950 | aspartate-alanine antiporter | - |
| BAU10_08080 | 1814111..1814323 | + | 213 | ANP64951 | hypothetical protein | - |
| BAU10_08085 | 1814334..1815197 | + | 864 | ANP64952 | efflux transporter periplasmic adaptor subunit | - |
| BAU10_08090 | 1815211..1816566 | + | 1356 | ANP64953 | fusaric acid resistance protein | - |
| BAU10_08095 | 1816868..1818523 | + | 1656 | ANP64954 | hypothetical protein | - |
| BAU10_08100 | 1818734..1820773 | + | 2040 | ANP64955 | para-nitrobenzyl esterase | - |
| BAU10_08105 | 1820875..1822326 | + | 1452 | ANP64956 | phospholipase | - |
| BAU10_08110 | 1822384..1824090 | - | 1707 | ANP64957 | hypothetical protein | - |
| BAU10_08115 | 1824378..1824842 | - | 465 | ANP64958 | hypothetical protein | - |
| BAU10_08120 | 1825012..1825638 | - | 627 | ANP64959 | lysine transporter LysE | - |
| BAU10_08125 | 1825966..1826592 | - | 627 | ANP64960 | amino acid transporter | - |
| BAU10_08130 | 1826722..1827075 | - | 354 | ANP64961 | RNA signal recognition particle | - |
| BAU10_08135 | 1827173..1827778 | - | 606 | ANP64962 | hypothetical protein | - |
| BAU10_08140 | 1827860..1828156 | - | 297 | ANP64963 | hypothetical protein | - |
| BAU10_08145 | 1828300..1828587 | - | 288 | ANP64964 | antibiotic biosynthesis monooxygenase | - |
| BAU10_08150 | 1828716..1829357 | - | 642 | ANP64965 | alanyl-tRNA editing protein | - |
| BAU10_08155 | 1829500..1830045 | - | 546 | ANP64966 | GCN5 family acetyltransferase | - |
| BAU10_08160 | 1830299..1830862 | - | 564 | ANP64967 | maltose O-acetyltransferase | - |
| BAU10_08165 | 1831006..1831419 | - | 414 | ANP64968 | hypothetical protein | - |
| BAU10_08170 | 1831561..1832169 | - | 609 | ANP64969 | phosphohydrolase | - |
| BAU10_08175 | 1832313..1832714 | - | 402 | ANP64970 | phosphoenolpyruvate synthase | - |
| BAU10_08180 | 1832819..1833340 | - | 522 | ANP64971 | acetyltransferase | - |
| BAU10_08185 | 1833680..1834636 | + | 957 | ANP64972 | transposase | - |
| BAU10_08190 | 1834659..1835135 | - | 477 | Protein_1598 | histone acetyltransferase | - |
| BAU10_08195 | 1835277..1835810 | - | 534 | ANP64973 | hypothetical protein | - |
| BAU10_08200 | 1836010..1837563 | - | 1554 | ANP66471 | hypothetical protein | - |
| BAU10_08205 | 1837993..1838949 | + | 957 | ANP64974 | transposase | - |
| BAU10_08210 | 1840224..1840424 | - | 201 | Protein_1602 | hypothetical protein | - |
| BAU10_08215 | 1840886..1841842 | + | 957 | ANP64975 | transposase | - |
| BAU10_08220 | 1843117..1843317 | - | 201 | Protein_1604 | hypothetical protein | - |
| BAU10_08225 | 1843779..1844735 | + | 957 | ANP64976 | transposase | - |
| BAU10_08230 | 1844758..1845288 | - | 531 | ANP64977 | hypothetical protein | - |
| BAU10_08235 | 1845432..1845836 | - | 405 | ANP64978 | hypothetical protein | - |
| BAU10_08240 | 1845990..1846568 | - | 579 | ANP64979 | hypothetical protein | - |
| BAU10_08245 | 1846908..1847864 | + | 957 | ANP64980 | transposase | - |
| BAU10_08250 | 1847899..1848351 | - | 453 | ANP64981 | hypothetical protein | - |
| BAU10_08255 | 1848458..1849063 | - | 606 | ANP64982 | hypothetical protein | - |
| BAU10_08260 | 1849260..1849544 | + | 285 | ANP64983 | damage-inducible protein J | - |
| BAU10_08265 | 1849541..1849819 | + | 279 | ANP64984 | plasmid stabilization protein ParE | - |
| BAU10_08270 | 1850572..1850784 | - | 213 | ANP64985 | hypothetical protein | - |
| BAU10_08275 | 1852023..1852742 | - | 720 | ANP64986 | hypothetical protein | - |
| BAU10_08280 | 1852890..1853540 | - | 651 | ANP64987 | hypothetical protein | - |
| BAU10_08285 | 1853884..1854840 | + | 957 | ANP64988 | transposase | - |
| BAU10_08290 | 1854875..1855342 | - | 468 | ANP64989 | XRE family transcriptional regulator | - |
| BAU10_08295 | 1855582..1855989 | - | 408 | ANP64990 | hypothetical protein | - |
| BAU10_08300 | 1856317..1857273 | + | 957 | ANP64991 | integrase | - |
| BAU10_08305 | 1858644..1859021 | + | 378 | ANP64992 | hypothetical protein | - |
| BAU10_08310 | 1859168..1862050 | - | 2883 | ANP64993 | hypothetical protein | - |
| BAU10_08315 | 1862025..1862447 | - | 423 | ANP64994 | hypothetical protein | - |
| BAU10_08320 | 1862466..1862954 | - | 489 | ANP64995 | hypothetical protein | - |
| BAU10_08325 | 1863048..1866410 | - | 3363 | ANP64996 | hypothetical protein | - |
| BAU10_08330 | 1866410..1866883 | - | 474 | ANP64997 | hypothetical protein | - |
| BAU10_08335 | 1866873..1869059 | - | 2187 | ANP64998 | hypothetical protein | - |
| BAU10_08340 | 1869310..1869660 | - | 351 | ANP64999 | hypothetical protein | - |
| BAU10_08345 | 1869677..1870123 | - | 447 | ANP65000 | hypothetical protein | - |
| BAU10_08350 | 1870139..1870525 | - | 387 | ANP65001 | hypothetical protein | - |
| BAU10_08355 | 1870522..1870959 | - | 438 | ANP65002 | hypothetical protein | - |
| BAU10_08360 | 1870962..1871312 | - | 351 | ANP65003 | hypothetical protein | - |
| BAU10_08365 | 1871319..1871651 | - | 333 | ANP65004 | hypothetical protein | - |
| BAU10_08370 | 1871644..1871856 | - | 213 | ANP65005 | hypothetical protein | - |
| BAU10_08375 | 1871917..1873113 | - | 1197 | ANP65006 | hypothetical protein | - |
| BAU10_08380 | 1873146..1873760 | - | 615 | ANP65007 | peptidase | - |
| BAU10_08385 | 1873753..1874952 | - | 1200 | ANP65008 | phage portal protein | - |
| BAU10_08390 | 1875103..1876836 | - | 1734 | ANP65009 | terminase | - |
| BAU10_08395 | 1876839..1877291 | - | 453 | ANP65010 | hypothetical protein | - |
| BAU10_08400 | 1877650..1878009 | - | 360 | ANP65011 | DNAse | - |
| BAU10_08405 | 1878348..1878590 | - | 243 | ANP65012 | hypothetical protein | - |
| BAU10_08410 | 1878702..1878965 | - | 264 | ANP65013 | hypothetical protein | - |
| BAU10_08415 | 1879006..1879332 | - | 327 | ANP66472 | hypothetical protein | - |
| BAU10_08420 | 1879346..1879636 | - | 291 | ANP65014 | hypothetical protein | - |
| BAU10_08425 | 1879707..1880396 | - | 690 | ANP65015 | hypothetical protein | - |
| BAU10_08430 | 1880502..1880876 | + | 375 | ANP65016 | hypothetical protein | - |
| BAU10_08435 | 1881073..1881678 | + | 606 | ANP65017 | hypothetical protein | - |
| BAU10_08440 | 1881888..1882136 | + | 249 | ANP65018 | hypothetical protein | - |
| BAU10_08445 | 1882172..1882450 | + | 279 | ANP65019 | hypothetical protein | - |
| BAU10_08450 | 1882562..1883533 | - | 972 | ANP65020 | hypothetical protein | - |
| BAU10_08455 | 1883544..1883849 | - | 306 | ANP65021 | nucleoside 2-deoxyribosyltransferase | - |
| BAU10_08460 | 1883846..1884154 | - | 309 | ANP65022 | hypothetical protein | - |
| BAU10_08465 | 1884151..1884339 | - | 189 | ANP65023 | hypothetical protein | - |
| BAU10_08470 | 1884336..1885037 | - | 702 | ANP65024 | DNA-binding protein | - |
| BAU10_08475 | 1885204..1886190 | - | 987 | ANP65025 | transcriptional regulator | - |
| BAU10_08480 | 1886190..1886768 | - | 579 | ANP65026 | phage N-6-adenine-methyltransferase | - |
| BAU10_08485 | 1887121..1887312 | - | 192 | ANP65027 | hypothetical protein | - |
| BAU10_08490 | 1887309..1887818 | - | 510 | ANP65028 | hypothetical protein | - |
| BAU10_08495 | 1888098..1888898 | + | 801 | ANP65029 | XRE family transcriptional regulator | - |
| BAU10_08500 | 1888992..1889534 | + | 543 | ANP65030 | hypothetical protein | - |
| BAU10_08505 | 1889880..1890272 | + | 393 | ANP65031 | hypothetical protein | - |
| BAU10_08510 | 1890313..1891131 | + | 819 | ANP65032 | hypothetical protein | - |
| BAU10_08515 | 1891183..1891560 | + | 378 | ANP65033 | hypothetical protein | - |
| BAU10_08520 | 1891562..1891774 | + | 213 | ANP65034 | hypothetical protein | - |
| BAU10_08525 | 1891767..1892024 | + | 258 | ANP65035 | hypothetical protein | - |
| BAU10_08530 | 1892024..1892239 | + | 216 | ANP65036 | hypothetical protein | - |
| BAU10_08535 | 1892344..1892574 | + | 231 | ANP65037 | hypothetical protein | - |
| BAU10_08540 | 1892574..1893809 | + | 1236 | ANP65038 | integrase | - |
| BAU10_08545 | 1893957..1894586 | - | 630 | ANP65039 | hypothetical protein | - |
| BAU10_08550 | 1894963..1895436 | - | 474 | ANP65040 | hypothetical protein | - |
| BAU10_08555 | 1895450..1896037 | - | 588 | ANP65041 | phosphohydrolase | - |
| BAU10_08560 | 1896039..1896623 | - | 585 | ANP65042 | ribosomal-protein-alanine acetyltransferase | - |
| BAU10_08565 | 1896660..1898204 | - | 1545 | ANP65043 | DNA-binding transcriptional regulator TyrR | - |
| BAU10_08570 | 1898305..1899345 | - | 1041 | ANP65044 | TIGR01620 family protein | - |
| BAU10_08575 | 1899342..1900718 | - | 1377 | ANP65045 | nucleoside triphosphate hydrolase | - |
| BAU10_08580 | 1900975..1901160 | - | 186 | ANP65046 | hypothetical protein | - |
| BAU10_08585 | 1901335..1902852 | - | 1518 | ANP65047 | fumarate hydratase | - |
| BAU10_08590 | 1903123..1904487 | + | 1365 | ANP65048 | aminodeoxychorismate synthase, component I | - |
| BAU10_08595 | 1904848..1906548 | - | 1701 | ANP65049 | hybrid sensor histidine kinase/response regulator | - |
| BAU10_08600 | 1906556..1907095 | - | 540 | ANP65050 | guanylate cyclase | - |
| BAU10_08605 | 1907327..1907926 | + | 600 | ANP65051 | coenzyme A pyrophosphatase | - |
| BAU10_08610 | 1908272..1909525 | + | 1254 | ANP65052 | septum formation protein | - |
| BAU10_08615 | 1909623..1910984 | + | 1362 | ANP65053 | L-serine ammonia-lyase | - |
| BAU10_08620 | 1911196..1912911 | + | 1716 | ANP65054 | diguanylate cyclase | - |
| BAU10_08625 | 1913006..1913473 | + | 468 | ANP65055 | acetyltransferase | - |
| BAU10_08630 | 1913563..1913925 | - | 363 | ANP65056 | 6-carboxytetrahydropterin synthase QueD | - |
| BAU10_08635 | 1914219..1915844 | + | 1626 | ANP65057 | chemotaxis protein | - |
| BAU10_08640 | 1916034..1917434 | + | 1401 | ANP65058 | asparagine--tRNA ligase | - |
| BAU10_08645 | 1917526..1917750 | - | 225 | ANP65059 | hypothetical protein | - |
| BAU10_08650 | 1917849..1919240 | - | 1392 | ANP65060 | GntR family transcriptional regulator | - |
| BAU10_08655 | 1919500..1919730 | + | 231 | ANP65061 | hypothetical protein | - |
| BAU10_08660 | 1919976..1920275 | + | 300 | ANP65062 | hypothetical protein | - |
| BAU10_08665 | 1920608..1921798 | + | 1191 | ANP65063 | aromatic amino acid aminotransferase | - |
| BAU10_08670 | 1921888..1922529 | - | 642 | ANP65064 | hypothetical protein | - |
| BAU10_08675 | 1922844..1924733 | + | 1890 | ANP65065 | cyclic nucleotide-binding protein | - |
| BAU10_08680 | 1924745..1925461 | + | 717 | ANP66473 | DNA polymerase III subunit epsilon | - |
| BAU10_08685 | 1925533..1927416 | - | 1884 | ANP65066 | chemotaxis protein | - |
| BAU10_08690 | 1927648..1929219 | - | 1572 | ANP65067 | BCCT transporter | - |
| BAU10_08695 | 1929607..1930083 | + | 477 | ANP66474 | MarR family transcriptional regulator | - |
| BAU10_08700 | 1930124..1930762 | - | 639 | ANP65068 | DNA-binding response regulator | - |
| BAU10_08705 | 1930755..1932110 | - | 1356 | ANP65069 | histidine kinase | - |
| BAU10_08710 | 1932262..1933548 | - | 1287 | ANP65070 | C4-dicarboxylate ABC transporter | - |
| BAU10_08715 | 1933561..1934100 | - | 540 | ANP65071 | C4-dicarboxylate ABC transporter | - |
| BAU10_08720 | 1934216..1935307 | - | 1092 | ANP65072 | ABC transporter substrate-binding protein | - |
| BAU10_08725 | 1935722..1936126 | - | 405 | ANP65073 | hypothetical protein | - |
| BAU10_08730 | 1936400..1937065 | + | 666 | ANP65074 | hypothetical protein | - |
| BAU10_08735 | 1937084..1937740 | - | 657 | ANP65075 | hypothetical protein | - |
| BAU10_08740 | 1937870..1938418 | - | 549 | ANP65076 | hypothetical protein | - |
| BAU10_08745 | 1938585..1940126 | - | 1542 | ANP65077 | peptidase | - |
| BAU10_08750 | 1940280..1941362 | - | 1083 | ANP65078 | hypothetical protein | - |
| BAU10_08755 | 1941343..1942380 | - | 1038 | ANP65079 | iron-regulated protein A | - |
| BAU10_08760 | 1942391..1943773 | - | 1383 | ANP65080 | thiol oxidoreductase | - |
| BAU10_08765 | 1943866..1945128 | - | 1263 | ANP65081 | peptidase | - |
| BAU10_08770 | 1945303..1945899 | - | 597 | ANP66475 | GTP cyclohydrolase | - |
| BAU10_08775 | 1946171..1946356 | - | 186 | ANP65082 | hypothetical protein | - |
| BAU10_08780 | 1946494..1946904 | - | 411 | ANP65083 | cytochrome C nitrite reductase | - |
| BAU10_08785 | 1946891..1947424 | - | 534 | ANP65084 | thiol:disulfide interchange protein | - |
| BAU10_08790 | 1947402..1949309 | - | 1908 | ANP65085 | cytochrome c nitrate reductase biogenesis protein NrfE | - |
| BAU10_08795 | 1949319..1950290 | - | 972 | ANP65086 | cytochrome c nitrite reductase subunit NrfD | - |
| BAU10_08800 | 1950287..1950973 | - | 687 | ANP65087 | cytochrome c nitrite reductase Fe-S protein | - |
| BAU10_08805 | 1950975..1951577 | - | 603 | ANP65088 | cytochrome c nitrite reductase pentaheme subunit | - |
| BAU10_08810 | 1951742..1953169 | + | 1428 | ANP66476 | nitrite reductase (cytochrome; ammonia-forming) c552 subunit | - |
| BAU10_08815 | 1953288..1953842 | - | 555 | ANP65089 | hypothetical protein | - |
| BAU10_08820 | 1953885..1954508 | - | 624 | ANP65090 | hypothetical protein | - |
| BAU10_08825 | 1954632..1957343 | - | 2712 | ANP65091 | DNA gyrase subunit A | - |
| BAU10_08830 | 1957819..1958526 | + | 708 | ANP65092 | bifunctional 3-demethylubiquinol 3-O-methyltransferase/2-polyprenyl-6-hydroxyphenol methylase | - |
| BAU10_08835 | 1959028..1961310 | + | 2283 | ANP65093 | ribonucleoside-diphosphate reductase subunit alpha | - |
| BAU10_08840 | 1961394..1962527 | + | 1134 | ANP65094 | ribonucleotide-diphosphate reductase subunit beta | - |
| BAU10_08845 | 1962527..1962808 | + | 282 | ANP65095 | (2Fe-2S)-binding protein | - |
| BAU10_08850 | 1962905..1964137 | - | 1233 | ANP65096 | competence/damage-inducible protein A | - |
| BAU10_08855 | 1964329..1965231 | + | 903 | ANP65097 | ubiquitin--protein ligase | - |
| BAU10_08860 | 1965453..1965965 | + | 513 | ANP65098 | transcriptional regulator | - |
| BAU10_08865 | 1966043..1967176 | - | 1134 | ANP65099 | carboxynorspermidine decarboxylase | - |
| BAU10_08870 | 1967280..1968524 | - | 1245 | ANP65100 | saccharopine dehydrogenase | - |
| BAU10_08875 | 1968543..1971419 | - | 2877 | ANP65101 | aminotransferase class III | - |
| BAU10_08900 | 1973540..1974097 | - | 558 | ANP65102 | CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase | - |
| BAU10_08905 | 1974144..1975910 | - | 1767 | ANP65103 | excinuclease ABC subunit C | - |
| BAU10_08910 | 1975978..1976622 | - | 645 | ANP65104 | two-component system response regulator UvrY | - |
| BAU10_08915 | 1977050..1979413 | + | 2364 | ANP65105 | DNA polymerase II | - |
| BAU10_08920 | 1979410..1980177 | - | 768 | ANP65106 | nucleotidyltransferase | - |
| BAU10_08925 | 1980372..1980938 | + | 567 | ANP65107 | elongation factor P-like protein YeiP | - |
| BAU10_08930 | 1980941..1981261 | + | 321 | ANP65108 | hypothetical protein | - |
| BAU10_08935 | 1981449..1982387 | - | 939 | ANP65109 | 23S rRNA pseudouridylate synthase B | - |
| BAU10_08940 | 1982953..1983663 | + | 711 | ANP65110 | hcalcium-binding protein | - |
| BAU10_08945 | 1983757..1984377 | - | 621 | ANP65111 | threonylcarbamoyl-AMP synthase | - |
| BAU10_08950 | 1984437..1985294 | - | 858 | ANP65112 | phosphatase | - |
| BAU10_08955 | 1985390..1985515 | + | 126 | ANP65113 | Trp operon leader peptide | - |
| BAU10_08960 | 1985692..1987257 | + | 1566 | ANP65114 | anthranilate synthase component I | - |
| BAU10_08965 | 1987274..1987882 | + | 609 | ANP65115 | anthranilate synthase component II | - |
| BAU10_08970 | 1987920..1988930 | + | 1011 | ANP65116 | anthranilate phosphoribosyltransferase | - |
| BAU10_08975 | 1988944..1990383 | + | 1440 | ANP65117 | bifunctional indole-3-glycerol phosphate synthase/phosphoribosylanthranilate isomerase | - |
| BAU10_08980 | 1990448..1991638 | + | 1191 | ANP65118 | tryptophan synthase subunit beta | - |
| BAU10_08985 | 1991638..1992444 | + | 807 | ANP65119 | tryptophan synthase subunit alpha | - |
| BAU10_08990 | 1992641..1993336 | + | 696 | ANP65120 | transcriptional regulator | - |
| BAU10_08995 | 1993463..1995004 | - | 1542 | ANP65121 | long-chain-fatty-acid--CoA ligase | - |
| BAU10_09000 | 1995227..1995436 | - | 210 | ANP65122 | hypothetical protein | - |
| BAU10_09005 | 1995548..1996807 | + | 1260 | ANP65123 | amino acid:proton symporter | - |
| BAU10_09010 | 1996964..1998148 | + | 1185 | ANP65124 | acyl-homoserine-lactone synthase | - |
| BAU10_09015 | 1998141..2000681 | + | 2541 | ANP65125 | hybrid sensor histidine kinase/response regulator | - |
| BAU10_09020 | 2000793..2001035 | + | 243 | ANP65126 | hypothetical protein | - |
| BAU10_09025 | 2001234..2001797 | + | 564 | ANP65127 | septation protein A | - |
| BAU10_09030 | 2002023..2002418 | + | 396 | ANP65128 | acyl-CoA thioesterase | - |
| BAU10_09035 | 2002534..2002830 | + | 297 | ANP65129 | hypothetical protein | - |
| BAU10_09040 | 2002834..2003226 | + | 393 | ANP65130 | hypothetical protein | - |
| BAU10_09045 | 2003360..2005642 | - | 2283 | ANP65131 | 5-methyltetrahydropteroyltriglutamate-- homocysteine S-methyltransferase | - |
| BAU10_09050 | 2005924..2006835 | + | 912 | ANP65132 | transcriptional regulator | - |
| BAU10_09055 | 2006857..2007057 | - | 201 | ANP65133 | hypothetical protein | - |
| BAU10_09060 | 2007210..2007920 | - | 711 | ANP65134 | hypothetical protein | - |
| BAU10_09065 | 2008006..2010462 | - | 2457 | ANP65135 | diguanylate phosphodiesterase | - |
| BAU10_09070 | 2010888..2013221 | + | 2334 | ANP65136 | YgiQ family radical SAM protein | - |
| BAU10_09075 | 2013484..2016981 | + | 3498 | ANP65137 | hypothetical protein | - |
| BAU10_09080 | 2016985..2017932 | + | 948 | ANP65138 | hypothetical protein | - |
| BAU10_09085 | 2017936..2018881 | + | 946 | Protein_1773 | hypothetical protein | - |
| BAU10_09090 | 2018885..2019829 | + | 945 | ANP65139 | hypothetical protein | - |
| BAU10_09095 | 2019957..2020379 | - | 423 | ANP65140 | polysaccharide biosynthesis protein GtrA | - |
| BAU10_09100 | 2020384..2021790 | - | 1407 | ANP65141 | dolichyl-phosphate-mannose--protein mannosyltransferase | - |
| BAU10_09105 | 2021793..2022794 | - | 1002 | ANP66477 | glycosyl transferase family 2 | - |
| BAU10_09110 | 2022986..2023663 | + | 678 | ANP65142 | two-component system response regulator | - |
| BAU10_09115 | 2023660..2024946 | + | 1287 | ANP65143 | two-component sensor histidine kinase | - |
| BAU10_09120 | 2025056..2025952 | + | 897 | ANP66478 | 1-aminocyclopropane-1-carboxylate deaminase | - |
| BAU10_09125 | 2025992..2026534 | - | 543 | ANP65144 | hydrolase | - |
| BAU10_09130 | 2026685..2027545 | - | 861 | ANP65145 | LysR family transcriptional regulator | - |
| BAU10_09135 | 2027645..2028709 | + | 1065 | ANP65146 | multidrug transporter | - |
| BAU10_09140 | 2028719..2029738 | + | 1020 | ANP65147 | MFS transporter | - |
| BAU10_09145 | 2030936..2031964 | - | 1029 | ANP65148 | 5-methyltetrahydropteroyltriglutamate-- homocysteine methyltransferase | - |
| BAU10_09150 | 2031995..2032969 | - | 975 | ANP65149 | hypothetical protein | - |
| BAU10_09155 | 2034070..2034987 | + | 918 | ANP65150 | dTDP-glucose 4,6-dehydratase | - |
| BAU10_09160 | 2035063..2035746 | + | 684 | ANP65151 | topoisomerase I | - |
| BAU10_09165 | 2035812..2036546 | - | 735 | ANP65152 | macrolide ABC transporter ATP-binding protein | - |
| BAU10_09170 | 2036539..2037822 | - | 1284 | ANP65153 | peptide ABC transporter permease | - |
| BAU10_09175 | 2037825..2039039 | - | 1215 | ANP65154 | ABC transporter substrate-binding protein | - |
| BAU10_09180 | 2039036..2040307 | - | 1272 | ANP65155 | hypothetical protein | - |
| BAU10_09185 | 2040300..2041433 | - | 1134 | ANP65156 | efflux transporter periplasmic adaptor subunit | - |
| BAU10_09190 | 2041662..2042066 | - | 405 | ANP66479 | hypothetical protein | - |
| BAU10_09195 | 2042099..2042872 | - | 774 | ANP65157 | hypothetical protein | - |
| BAU10_09200 | 2043296..2043517 | + | 222 | ANP65158 | hypothetical protein | - |
| BAU10_09205 | 2043567..2044082 | - | 516 | ANP65159 | redoxin | - |
| BAU10_09210 | 2044079..2046190 | - | 2112 | ANP65160 | cytochrome C biogenesis protein | - |
| BAU10_09215 | 2046255..2046653 | - | 399 | ANP65161 | hypothetical protein | - |
| BAU10_09220 | 2046756..2047358 | - | 603 | ANP65162 | DNA-binding response regulator | - |
| BAU10_09225 | 2047384..2049210 | - | 1827 | ANP65163 | histidine kinase | - |
| BAU10_09230 | 2049390..2050154 | + | 765 | ANP65164 | 4Fe-4S ferredoxin | - |
| BAU10_09235 | 2050151..2051281 | + | 1131 | ANP65165 | tetrathionate reductase | - |
| BAU10_09240 | 2051299..2054391 | + | 3093 | ANP65166 | tetrathionate reductase subunit A | - |
| BAU10_09245 | 2054621..2055232 | + | 612 | ANP65167 | cystathionine beta-synthase | - |
| BAU10_09250 | 2055241..2056293 | + | 1053 | ANP65168 | cytochrome C | - |
| BAU10_09255 | 2056598..2057929 | + | 1332 | ANP65169 | paraquat-inducible protein A | - |
| BAU10_09260 | 2057922..2059568 | + | 1647 | ANP65170 | paraquat-inducible protein B | - |
| BAU10_09265 | 2059568..2060146 | + | 579 | ANP65171 | hypothetical protein | - |
| BAU10_09270 | 2060249..2061022 | + | 774 | ANP65172 | hypothetical protein | - |
| BAU10_09275 | 2061229..2061468 | + | 240 | ANP65173 | hypothetical protein | - |
| BAU10_09280 | 2061529..2063367 | - | 1839 | ANP65174 | diguanylate cyclase | - |
| BAU10_09285 | 2063867..2064568 | - | 702 | ANP65175 | orotidine 5'-phosphate decarboxylase | - |
| BAU10_09290 | 2064653..2065828 | - | 1176 | ANP65176 | lipopolysaccharide assembly protein LapB | - |
| BAU10_09295 | 2065847..2066143 | - | 297 | ANP65177 | hypothetical protein | - |
| BAU10_09300 | 2066413..2066694 | - | 282 | ANP65178 | integration host factor subunit beta | - |
| BAU10_09305 | 2066908..2068578 | - | 1671 | ANP65179 | 30S ribosomal protein S1 | - |
| BAU10_09310 | 2068689..2069369 | - | 681 | ANP66480 | cytidylate kinase | - |
| BAU10_09315 | 2069578..2070639 | - | 1062 | ANP65180 | protease SohB | - |
| BAU10_09320 | 2070894..2071640 | + | 747 | ANP65181 | YciK family oxidoreductase | - |
| BAU10_09325 | 2071770..2072462 | + | 693 | ANP65182 | recombinase RecA | - |
| BAU10_09330 | 2072467..2073870 | + | 1404 | ANP65183 | nucleotidyltransferase | - |
| BAU10_09335 | 2073898..2076972 | + | 3075 | ANP65184 | error-prone DNA polymerase | - |
| BAU10_09340 | 2077045..2077998 | - | 954 | ANP65185 | chemotaxis protein CheW | - |
| BAU10_09345 | 2078362..2079579 | - | 1218 | ANP65186 | transcriptional regulator | - |
| BAU10_09350 | 2079990..2081432 | + | 1443 | ANP65187 | pyruvate kinase | - |
| BAU10_09355 | 2081541..2082692 | - | 1152 | ANP65188 | patatin family protein | - |
| BAU10_09360 | 2082957..2083547 | + | 591 | ANP65189 | hypothetical protein | - |
| BAU10_09365 | 2084117..2085583 | + | 1467 | ANP65190 | transporter | - |
| BAU10_09370 | 2085744..2086145 | - | 402 | ANP65191 | hypothetical protein | - |
| BAU10_09375 | 2086359..2087789 | - | 1431 | ANP65192 | PTS glucose transporter subunit IIBC | - |
| BAU10_09380 | 2088234..2089001 | - | 768 | ANP65193 | metal-dependent hydrolase | - |
| BAU10_09385 | 2088992..2089954 | - | 963 | ANP65194 | DNA polymerase III subunit delta' | - |
| BAU10_09390 | 2089979..2090611 | - | 633 | ANP65195 | dTMP kinase | - |
| BAU10_09395 | 2090608..2091624 | - | 1017 | ANP65196 | ABC transporter substrate-binding protein | - |
| BAU10_09400 | 2091621..2092436 | - | 816 | ANP65197 | aminodeoxychorismate lyase | - |
| BAU10_09405 | 2092544..2093794 | - | 1251 | ANP65198 | beta-ketoacyl-[acyl-carrier-protein] synthase II | - |
| BAU10_09410 | 2093888..2094121 | - | 234 | ANP65199 | acyl carrier protein | - |
| BAU10_09415 | 2094368..2095102 | - | 735 | ANP66481 | 3-oxoacyl-ACP reductase | - |
| BAU10_09420 | 2095148..2096071 | - | 924 | ANP65200 | malonyl CoA-acyl carrier protein transacylase | - |
| BAU10_09425 | 2096147..2097097 | - | 951 | ANP65201 | 3-oxoacyl-ACP synthase | - |
| BAU10_09430 | 2097103..2098128 | - | 1026 | ANP65202 | phosphate acyltransferase | - |
| BAU10_09435 | 2098139..2098309 | - | 171 | ANP65203 | 50S ribosomal protein L32 | - |
| BAU10_09440 | 2098369..2098893 | - | 525 | ANP65204 | ribosomal protein L32p | - |
| BAU10_09445 | 2099043..2099624 | + | 582 | ANP65205 | septum formation protein Maf | - |
| BAU10_09450 | 2099683..2100627 | - | 945 | ANP66482 | RNA pseudouridine synthase | - |
| BAU10_09455 | 2101267..2104332 | + | 3066 | ANP65206 | ribonuclease | - |
| BAU10_09460 | 2104665..2106224 | + | 1560 | ANP65207 | sodium-independent anion transporter | - |
| BAU10_09465 | 2106285..2106734 | - | 450 | ANP65208 | protein-tyrosine-phosphatase | - |
| BAU10_09470 | 2106852..2107457 | + | 606 | ANP65209 | cob(I)yrinic acid a,c-diamide adenosyltransferase | - |
| BAU10_09475 | 2107520..2109607 | - | 2088 | ANP65210 | cell envelope biogenesis protein AsmA | - |
| BAU10_09480 | 2109796..2110437 | - | 642 | ANP65211 | uridine kinase | - |
| BAU10_09485 | 2110618..2111694 | - | 1077 | ANP65212 | sodium:proton antiporter | - |
| BAU10_09490 | 2111857..2113932 | + | 2076 | ANP65213 | methionine--tRNA ligase | - |
| BAU10_09495 | 2114143..2114952 | + | 810 | ANP65214 | hypothetical protein | - |
| BAU10_09500 | 2115063..2115902 | - | 840 | ANP65215 | fatty acid metabolism regulator | - |
| BAU10_09505 | 2116542..2118128 | + | 1587 | ANP65216 | Na+/H+ antiporter NhaB | - |
| BAU10_09510 | 2118212..2118748 | + | 537 | ANP65217 | disulfide bond formation protein B | - |
| BAU10_09515 | 2118837..2119445 | - | 609 | ANP65218 | prepilin peptidase | - |
| BAU10_09520 | 2119616..2120575 | + | 960 | ANP65219 | tRNA dihydrouridine synthase DusC | - |
| BAU10_09525 | 2120627..2122087 | - | 1461 | ANP65220 | diguanylate cyclase | - |
| BAU10_09530 | 2122262..2124088 | - | 1827 | ANP65221 | maltodextrin glucosidase | - |
| BAU10_09535 | 2124230..2125024 | - | 795 | ANP65222 | ABC transporter ATP-binding protein | - |
| BAU10_09540 | 2125021..2125956 | - | 936 | ANP65223 | ABC transporter permease | - |
| BAU10_09545 | 2126132..2127097 | - | 966 | ANP65224 | ABC transporter substrate-binding protein | - |
| BAU10_09550 | 2127428..2127880 | - | 453 | ANP65225 | hypothetical protein | - |
| BAU10_09555 | 2128227..2129423 | + | 1197 | ANP65226 | acetate kinase | - |
| BAU10_09560 | 2129560..2131704 | + | 2145 | ANP65227 | phosphate acetyltransferase | - |
| BAU10_09565 | 2132047..2132469 | + | 423 | ANP65228 | hypothetical protein | - |
| BAU10_09570 | 2132583..2133530 | - | 948 | ANP65229 | glutathione-dependent reductase | - |
| BAU10_09575 | 2133698..2134711 | - | 1014 | ANP65230 | oligopeptide ABC transporter ATP-binding protein OppF | - |
| BAU10_09580 | 2134690..2135661 | - | 972 | ANP65231 | oligopeptide ABC transporter ATP-binding protein OppD | - |
| BAU10_09585 | 2135689..2136591 | - | 903 | ANP65232 | peptide ABC transporter permease | - |
| BAU10_09590 | 2136607..2137527 | - | 921 | ANP65233 | oligopeptide transporter permease | - |
| BAU10_09595 | 2137656..2139332 | - | 1677 | ANP66483 | oligopeptide ABC transporter substrate-binding protein OppA | - |
| BAU10_09600 | 2139864..2140319 | - | 456 | ANP65234 | molybdopterin synthase catalytic subunit | - |
| BAU10_09605 | 2140321..2140578 | - | 258 | ANP65235 | molybdopterin synthase sulfur carrier subunit | - |
| BAU10_09610 | 2140575..2141054 | - | 480 | ANP65236 | molybdenum cofactor biosynthesis protein C | - |
| BAU10_09615 | 2141081..2141593 | - | 513 | ANP65237 | molybdenum cofactor biosynthesis protein B | - |
| BAU10_09620 | 2141695..2142684 | - | 990 | ANP65238 | cyclic pyranopterin phosphate synthase | - |
| BAU10_09625 | 2142983..2143879 | + | 897 | ANP65239 | hypothetical protein | - |
| BAU10_09630 | 2144018..2144362 | - | 345 | ANP65240 | phosphorelay protein LuxU | - |
| BAU10_09635 | 2144359..2145765 | - | 1407 | ANP65241 | sigma-54-dependent Fis family transcriptional regulator | - |
| BAU10_09640 | 2146056..2148086 | - | 2031 | ANP65242 | excinuclease ABC subunit B | - |
| BAU10_09650 | 2149071..2151368 | + | 2298 | ANP65243 | formate acetyltransferase | - |
| BAU10_09655 | 2151674..2152252 | + | 579 | ANP65244 | electron transport complex subunit RsxA | - |
| BAU10_09660 | 2152256..2152852 | + | 597 | ANP65245 | electron transport complex subunit RsxB | - |
| BAU10_09665 | 2152861..2155269 | + | 2409 | ANP65246 | electron transport complex subunit RsxC | - |
| BAU10_09670 | 2155269..2156315 | + | 1047 | ANP65247 | electron transport complex subunit RsxD | - |
| BAU10_09675 | 2156323..2156955 | + | 633 | ANP65248 | electron transport complex subunit RsxG | - |
| BAU10_09680 | 2156955..2157647 | + | 693 | ANP65249 | electron transport complex subunit RsxE | - |
| BAU10_09685 | 2157712..2158353 | + | 642 | ANP65250 | endonuclease III | - |
| BAU10_09690 | 2158386..2158802 | + | 417 | ANP65251 | lactoylglutathione lyase | - |
| BAU10_09695 | 2158953..2159387 | + | 435 | ANP65252 | hypothetical protein | - |
| BAU10_09700 | 2159508..2160389 | - | 882 | ANP65253 | flagellar protein MotY | - |
| BAU10_09705 | 2160731..2161375 | + | 645 | ANP65254 | ribonuclease T | - |
| BAU10_09710 | 2161404..2162729 | + | 1326 | ANP65255 | sodium:proton antiporter | - |
| BAU10_09715 | 2162807..2163436 | - | 630 | ANP65256 | thioredoxin | - |
| BAU10_09720 | 2163573..2164148 | - | 576 | ANP65257 | hypothetical protein | - |
| BAU10_09725 | 2164342..2164917 | - | 576 | ANP65258 | hypothetical protein | - |
| BAU10_09730 | 2165335..2165667 | - | 333 | ANP65259 | monothiol glutaredoxin, Grx4 family | - |
| BAU10_09735 | 2165952..2166536 | + | 585 | ANP65260 | superoxide dismutase | - |
| BAU10_09740 | 2166691..2167194 | + | 504 | ANP65261 | hypothetical protein | - |
| BAU10_09745 | 2167329..2168072 | + | 744 | ANP65262 | short-chain dehydrogenase | - |
| BAU10_09750 | 2168217..2170919 | - | 2703 | ANP65263 | bifunctional acetaldehyde-CoA/alcohol dehydrogenase | - |
| BAU10_09755 | 2171570..2172208 | + | 639 | ANP65264 | hypothetical protein | - |
| BAU10_09760 | 2172259..2173107 | + | 849 | ANP66484 | ion transporter | - |
| BAU10_09765 | 2173165..2174280 | - | 1116 | ANP65265 | aspartate-semialdehyde dehydrogenase | - |
| BAU10_09770 | 2174712..2176145 | + | 1434 | ANP65266 | Na+/H+ antiporter NhaC | - |
| BAU10_09775 | 2176236..2177399 | - | 1164 | ANP65267 | chemotaxis protein | - |
| BAU10_09780 | 2177504..2178808 | - | 1305 | ANP66485 | histidine phosphotransferase | - |
| BAU10_09785 | 2178915..2179913 | - | 999 | ANP65268 | nucleoid-associated protein YejK | - |
| BAU10_09790 | 2179999..2180226 | + | 228 | ANP65269 | hypothetical protein | - |
| BAU10_09795 | 2180249..2182057 | + | 1809 | ANP65270 | hydrolase | - |
| BAU10_09805 | 2182498..2183421 | - | 924 | ANP65271 | histone deacetylase | - |
| BAU10_09810 | 2183693..2184382 | + | 690 | ANP65272 | short-chain dehydrogenase | - |
| BAU10_09815 | 2184566..2184772 | + | 207 | ANP65273 | hypothetical protein | - |
| BAU10_09820 | 2184817..2186784 | - | 1968 | ANP65274 | DNA topoisomerase III | - |
| BAU10_09825 | 2186888..2187436 | - | 549 | ANP65275 | nitroreductase | - |
| BAU10_09830 | 2187610..2188926 | + | 1317 | ANP65276 | MFS transporter | - |
| BAU10_09835 | 2188986..2190998 | - | 2013 | ANP65277 | NADPH-dependent 2,4-dienoyl-CoA reductase | - |
| BAU10_09840 | 2191223..2193073 | + | 1851 | ANP65278 | signal peptide peptidase SppA | - |
| BAU10_09845 | 2193238..2194251 | + | 1014 | ANP65279 | L-asparaginase 1 | - |
| BAU10_09850 | 2194347..2194631 | - | 285 | ANP65280 | hypothetical protein | - |
| BAU10_09855 | 2194733..2195554 | + | 822 | ANP65281 | hypothetical protein | - |
| BAU10_09860 | 2195673..2196080 | - | 408 | ANP65282 | peptide-methionine (R)-S-oxide reductase | - |
| BAU10_09865 | 2196418..2197413 | + | 996 | ANP65283 | type I glyceraldehyde-3-phosphate dehydrogenase | - |
| BAU10_09870 | 2197570..2198454 | + | 885 | ANP65284 | D-hexose-6-phosphate mutarotase | - |
| BAU10_09875 | 2198653..2200689 | + | 2037 | ANP65285 | chemotaxis protein | - |
| BAU10_09880 | 2200786..2201613 | + | 828 | ANP65286 | 23S rRNA (guanine(745)-N(1))-methyltransferase | - |
| BAU10_09885 | 2201712..2202659 | + | 948 | ANP65287 | hypothetical protein | - |
| BAU10_09890 | 2202911..2203270 | + | 360 | ANP65288 | hypothetical protein | - |
| BAU10_09895 | 2203669..2205246 | + | 1578 | ANP65289 | alkaline phosphatase | - |
| BAU10_09900 | 2205383..2205682 | + | 300 | ANP65290 | aryl-sulfate sulfotransferase | - |
| BAU10_09905 | 2205823..2206218 | + | 396 | ANP65291 | lactoylglutathione lyase | - |
| BAU10_09910 | 2206236..2206520 | + | 285 | ANP65292 | antibiotic biosynthesis monooxygenase | - |
| BAU10_09915 | 2206596..2208275 | - | 1680 | ANP65293 | mechanosensitive ion channel protein MscS | - |
| BAU10_09920 | 2208467..2208907 | - | 441 | ANP65294 | hypothetical protein | - |
| BAU10_09925 | 2209096..2209344 | + | 249 | ANP65295 | hypothetical protein | - |
| BAU10_09930 | 2209959..2210657 | + | 699 | ANP65296 | aquaporin Z | - |
| BAU10_09935 | 2210709..2211308 | - | 600 | ANP65297 | recombination protein RecR | - |
| BAU10_09940 | 2211339..2211668 | - | 330 | ANP65298 | nucleoid-associated protein | - |
| BAU10_09945 | 2211751..2213856 | - | 2106 | ANP65299 | DNA polymerase III subunit gamma/tau | - |
| BAU10_09950 | 2213865..2214410 | - | 546 | ANP65300 | adenine phosphoribosyltransferase | - |
| BAU10_09955 | 2214573..2214920 | - | 348 | ANP65301 | hypothetical protein | - |
| BAU10_09960 | 2215348..2216454 | - | 1107 | ANP65302 | two-component system response regulator | - |
| BAU10_09965 | 2216744..2217619 | + | 876 | ANP65303 | LysR family transcriptional regulator | - |
| BAU10_09970 | 2217685..2219199 | - | 1515 | ANP65304 | amidophosphoribosyltransferase | - |
| BAU10_09975 | 2219236..2219727 | - | 492 | ANP65305 | bacteriocin production protein | - |
| BAU10_09980 | 2219789..2220382 | - | 594 | ANP65306 | cell division protein DedD | - |
| BAU10_09985 | 2220397..2221659 | - | 1263 | ANP65307 | bifunctional folylpolyglutamate synthase/dihydrofolate synthase | - |
| BAU10_09990 | 2221706..2222632 | - | 927 | ANP65308 | acetyl-CoA carboxylase subunit beta | - |
| BAU10_09995 | 2222830..2223624 | - | 795 | ANP65309 | tRNA pseudouridine(38,39,40) synthase TruA | - |
| BAU10_10000 | 2223767..2227918 | - | 4152 | ANP65310 | AAA family ATPase | - |
| BAU10_10005 | 2228116..2229129 | - | 1014 | ANP65311 | aspartate-semialdehyde dehydrogenase | - |
| BAU10_10010 | 2229126..2230259 | - | 1134 | ANP65312 | 4-phosphoerythronate dehydrogenase | - |
| BAU10_10015 | 2230325..2230603 | + | 279 | ANP65313 | erythronate-4-phosphate dehydrogenase | - |
| BAU10_10020 | 2230600..2231811 | - | 1212 | ANP65314 | beta-ketoacyl-[acyl-carrier-protein] synthase I | - |
| BAU10_10025 | 2231940..2233958 | + | 2019 | ANP65315 | bifunctional tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase MnmD/FAD-dependent cmnm(5)s(2)U34 oxidoreductase MnmC | - |
| BAU10_10030 | 2234035..2234559 | - | 525 | ANP65316 | NADPH-dependent FMN reductase | - |
| BAU10_10035 | 2234706..2234969 | - | 264 | ANP65317 | hypothetical protein | - |
| BAU10_10040 | 2234995..2235525 | - | 531 | ANP65318 | elongation factor P hydroxylase | - |
| BAU10_10045 | 2235858..2236481 | + | 624 | ANP65319 | hypothetical protein | - |
| BAU10_10050 | 2236705..2237790 | - | 1086 | ANP65320 | chorismate synthase | - |
| BAU10_10055 | 2237971..2238903 | - | 933 | ANP65321 | ribosomal protein L3 N(5)-glutamine methyltransferase | - |
| BAU10_10060 | 2238966..2239496 | + | 531 | ANP65322 | hypothetical protein | - |
| BAU10_10065 | 2239593..2240057 | - | 465 | ANP65323 | phosphohistidine phosphatase SixA | - |
| BAU10_10070 | 2240432..2243209 | + | 2778 | ANP65324 | peptidase M16 | - |
| BAU10_10075 | 2243376..2243630 | + | 255 | ANP65325 | CG2 omega domain protein | - |
| BAU10_10080 | 2243693..2245804 | - | 2112 | ANP65326 | multifunctional fatty acid oxidation complex subunit alpha | - |
| BAU10_10085 | 2245804..2247111 | - | 1308 | ANP65327 | 3-ketoacyl-CoA thiolase | - |
| BAU10_10090 | 2247403..2247633 | - | 231 | ANP65328 | acetyl-CoA acetyltransferase | - |
| BAU10_10095 | 2247832..2248407 | + | 576 | ANP65329 | RNA polymerase subunit sigma | - |
| BAU10_10100 | 2248400..2249131 | + | 732 | ANP65330 | chemotaxis protein | - |
| BAU10_10105 | 2249429..2250706 | + | 1278 | ANP65331 | long-chain fatty acid transporter | - |
| BAU10_10110 | 2251016..2252332 | + | 1317 | ANP65332 | aromatic hydrocarbon degradation protein | - |
| BAU10_10115 | 2252407..2253192 | - | 786 | ANP65333 | ABC transporter | - |
| BAU10_10120 | 2253397..2254611 | - | 1215 | ANP65334 | c-type cytochrome biogenesis protein CcmI | - |
| BAU10_10125 | 2254611..2255096 | - | 486 | ANP65335 | cystathionine gamma-synthase | - |
| BAU10_10130 | 2255093..2255647 | - | 555 | ANP65336 | thiol:disulfide interchange protein | - |
| BAU10_10135 | 2255649..2257625 | - | 1977 | ANP65337 | c-type cytochrome biogenesis protein CcmF | - |
| BAU10_10140 | 2257622..2258107 | - | 486 | ANP65338 | cytochrome c biogenesis protein CcmE | - |
| BAU10_10145 | 2258104..2258310 | - | 207 | ANP65339 | heme exporter protein CcmD | - |
| BAU10_10150 | 2258322..2259065 | - | 744 | ANP65340 | heme ABC transporter permease | - |
| BAU10_10155 | 2259175..2259843 | - | 669 | ANP65341 | heme exporter protein CcmB | - |
| BAU10_10160 | 2259845..2260462 | - | 618 | ANP65342 | heme ABC transporter ATP-binding protein CcmA | - |
| BAU10_10165 | 2260752..2261246 | - | 495 | ANP66486 | DNA repair protein | - |
| BAU10_10170 | 2261260..2261754 | - | 495 | ANP65343 | chemotaxis protein CheW | - |
| BAU10_10175 | 2261790..2262842 | - | 1053 | ANP65344 | chemotaxis protein CheW | - |
| BAU10_10180 | 2262832..2263611 | - | 780 | ANP65345 | cobalamin biosynthesis protein CobQ | - |
| BAU10_10185 | 2263620..2264732 | - | 1113 | ANP65346 | chemotaxis response regulator protein-glutamate methylesterase | - |
| BAU10_10190 | 2264762..2267002 | - | 2241 | ANP65347 | chemotaxis protein CheA | - |
| BAU10_10195 | 2267012..2267752 | - | 741 | ANP65348 | protein phosphatase | - |
| BAU10_10200 | 2267871..2268239 | - | 369 | ANP65349 | histidine kinase | - |
| BAU10_10205 | 2268285..2269019 | - | 735 | ANP65350 | RNA polymerase sigma factor FliA | - |
| BAU10_10210 | 2269012..2269899 | - | 888 | ANP65351 | cobyrinic acid a,c-diamide synthase | - |
| BAU10_10215 | 2269914..2271431 | - | 1518 | ANP65352 | flagellar biosynthesis protein FlhF | - |
| BAU10_10220 | 2271460..2273559 | - | 2100 | ANP66487 | flagellar biosynthesis protein FlhA | - |
| BAU10_10225 | 2273770..2274900 | - | 1131 | ANP65353 | flagellar biosynthesis protein FlhB | - |
| BAU10_10230 | 2274904..2275686 | - | 783 | ANP65354 | flagellar biosynthetic protein FliR | - |
| BAU10_10235 | 2275695..2275964 | - | 270 | ANP65355 | flagellar export apparatus protein FliQ | - |
| BAU10_10240 | 2275989..2276858 | - | 870 | ANP65356 | flagellar biosynthetic protein FliP | - |
| BAU10_10245 | 2276845..2277258 | - | 414 | ANP65357 | flagellar biosynthetic protein FliO | - |
| BAU10_10250 | 2277260..2277670 | - | 411 | ANP65358 | flagellar motor switch protein FliN | - |
| BAU10_10255 | 2277729..2278775 | - | 1047 | ANP65359 | flagellar motor switch protein FliM | - |
| BAU10_10260 | 2278783..2279286 | - | 504 | ANP65360 | flagellar basal body-associated protein FliL | - |
| BAU10_10265 | 2279372..2281225 | - | 1854 | ANP65361 | flagellar hook-length control protein FliK | - |
| BAU10_10270 | 2281506..2281820 | + | 315 | ANP65362 | hypothetical protein | - |
| BAU10_10275 | 2281861..2282304 | - | 444 | ANP65363 | flagellar protein FliJ | - |
| BAU10_10280 | 2282311..2283630 | - | 1320 | ANP65364 | flagellar protein export ATPase FliI | - |
| BAU10_10285 | 2283634..2284434 | - | 801 | ANP65365 | flagellar assembly protein FliH | - |
| BAU10_10290 | 2284506..2285561 | - | 1056 | ANP65366 | flagellar motor switch protein FliG | - |
| BAU10_10295 | 2285554..2287302 | - | 1749 | ANP65367 | flagellar M-ring protein FliF | - |
| BAU10_10300 | 2287311..2287622 | - | 312 | ANP66488 | flagellar hook-basal body complex protein FliE | - |
| BAU10_10305 | 2287745..2289154 | - | 1410 | ANP65368 | sigma-54-dependent Fis family transcriptional regulator | - |
| BAU10_10310 | 2289191..2290222 | - | 1032 | ANP65369 | PAS domain-containing sensor histidine kinase | - |
| BAU10_10315 | 2290422..2291888 | - | 1467 | ANP65370 | sigma-54-dependent Fis family transcriptional regulator | - |
| BAU10_10320 | 2292176..2292586 | - | 411 | ANP65371 | flagellar export chaperone FliS | - |
| BAU10_10325 | 2292599..2292904 | - | 306 | ANP65372 | flagellar rod protein FlaI | - |
| BAU10_10330 | 2292908..2294902 | - | 1995 | ANP65373 | flagellar filament capping protein FliD | - |
| BAU10_10335 | 2294924..2295358 | - | 435 | ANP65374 | flagellar biosynthesis protein FlaG | - |
| BAU10_10340 | 2295451..2296581 | - | 1131 | ANP65375 | flagellin B | - |
| BAU10_10345 | 2296870..2298003 | - | 1134 | ANP65376 | flagellin | - |
| BAU10_10350 | 2298266..2299405 | - | 1140 | ANP65377 | flagellin | - |
| BAU10_10355 | 2299549..2300451 | - | 903 | ANP65378 | deferrochelatase | - |
| BAU10_10360 | 2300498..2300983 | - | 486 | ANP66489 | hypothetical protein | - |
| BAU10_10365 | 2301128..2301481 | + | 354 | ANP65379 | hypothetical protein | - |
| BAU10_10370 | 2301485..2301817 | + | 333 | ANP65380 | hypothetical protein | - |
| BAU10_10375 | 2301852..2302139 | - | 288 | ANP65381 | hypothetical protein | - |
| BAU10_10380 | 2302431..2302781 | + | 351 | ANP65382 | arsenate reductase | - |
| BAU10_10385 | 2302800..2303936 | + | 1137 | ANP65383 | succinyl-diaminopimelate desuccinylase | - |
| BAU10_10390 | 2303951..2304649 | + | 699 | ANP65384 | peptidase M15 | - |
| BAU10_10395 | 2304646..2304849 | + | 204 | ANP65385 | DUF2897 domain-containing protein | - |
| BAU10_10400 | 2304956..2305975 | - | 1020 | ANP65386 | outer membrane assembly protein BamC | - |
| BAU10_10405 | 2306056..2306934 | - | 879 | ANP65387 | 4-hydroxy-tetrahydrodipicolinate synthase | - |
| BAU10_10410 | 2307410..2307952 | + | 543 | ANP65388 | glycine cleavage system transcriptional repressor | - |
| BAU10_10415 | 2307969..2308439 | + | 471 | ANP65389 | thioredoxin-dependent thiol peroxidase | - |
| BAU10_10420 | 2308517..2309590 | - | 1074 | ANP65390 | AI-2E family transporter | - |
| BAU10_10425 | 2309592..2309828 | - | 237 | ANP65391 | sirA-like family protein | - |
| BAU10_10430 | 2310086..2311537 | + | 1452 | ANP65392 | hypothetical protein | - |
| BAU10_10435 | 2311554..2311904 | + | 351 | ANP65393 | arsenate reductase (glutaredoxin) | - |
| BAU10_10440 | 2311905..2312474 | + | 570 | ANP65394 | NAD(P)H:quinone oxidoreductase, type IV | - |
| BAU10_10445 | 2312497..2312937 | + | 441 | ANP65395 | hypothetical protein | - |
| BAU10_10450 | 2313078..2314286 | - | 1209 | ANP65396 | hypothetical protein | - |
| BAU10_10455 | 2314652..2315905 | + | 1254 | ANP65397 | uracil permease | - |
| BAU10_10460 | 2316022..2316648 | - | 627 | ANP65398 | uracil phosphoribosyltransferase | - |
| BAU10_10465 | 2316828..2317868 | + | 1041 | ANP65399 | phosphoribosylformylglycinamidine cyclo-ligase | - |
| BAU10_10470 | 2318020..2318676 | + | 657 | ANP65400 | phosphoribosylglycinamide formyltransferase | - |
| BAU10_10475 | 2319066..2319911 | - | 846 | ANP65401 | class II glutamine amidotransferase | - |
| BAU10_10480 | 2319963..2320538 | - | 576 | ANP65402 | phosphoheptose isomerase | - |
| BAU10_10485 | 2320915..2323359 | + | 2445 | ANP65403 | acyl-CoA dehydrogenase | - |
| BAU10_10490 | 2323564..2325339 | + | 1776 | ANP65404 | sulfate permease | - |
| BAU10_10495 | 2325413..2325823 | - | 411 | ANP65405 | GCN5 family acetyltransferase | - |
| BAU10_10500 | 2325908..2326300 | - | 393 | ANP65406 | hypothetical protein | - |
| BAU10_10505 | 2326492..2327010 | - | 519 | ANP65407 | hypothetical protein | - |
| BAU10_10510 | 2327234..2328484 | - | 1251 | ANP65408 | TIGR03503 family protein | - |
| BAU10_10515 | 2328493..2329215 | - | 723 | ANP65409 | DNA polymerase III subunit epsilon | - |
| BAU10_10520 | 2329275..2329739 | + | 465 | ANP65410 | ribonuclease HI | - |
| BAU10_10525 | 2329736..2330482 | - | 747 | ANP65411 | SAM-dependent methyltransferase | - |
| BAU10_10530 | 2330520..2331278 | + | 759 | ANP66490 | hydroxyacylglutathione hydrolase | - |
| BAU10_10535 | 2331533..2333116 | + | 1584 | ANP65412 | lytic transglycosylase | - |
| BAU10_10540 | 2333113..2333799 | + | 687 | ANP65413 | hypothetical protein | - |
| BAU10_10545 | 2333801..2334646 | - | 846 | ANP65414 | hypothetical protein | - |
| BAU10_10550 | 2334787..2335125 | - | 339 | ANP65415 | transcriptional regulator | - |
| BAU10_10555 | 2335312..2335626 | - | 315 | ANP65416 | cytochrome C biogenesis protein CcsB | - |
| BAU10_10560 | 2335697..2337007 | - | 1311 | ANP65417 | tRNA lysidine(34) synthetase TilS | - |
| BAU10_10565 | 2337114..2338073 | - | 960 | ANP65418 | acetyl-CoA carboxylase carboxyltransferase subunit alpha | - |
| BAU10_10570 | 2338120..2341599 | - | 3480 | ANP65419 | DNA polymerase III subunit alpha | - |
| BAU10_10575 | 2341681..2342316 | - | 636 | ANP65420 | ribonuclease HII | - |
| BAU10_10580 | 2342326..2343465 | - | 1140 | ANP65421 | lipid-A-disaccharide synthase | - |
| BAU10_10585 | 2343585..2344373 | - | 789 | ANP65422 | acyl-[acyl-carrier-protein]--UDP-N- acetylglucosamine O-acyltransferase | - |
| BAU10_10590 | 2344375..2344827 | - | 453 | ANP65423 | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ | - |
| BAU10_10595 | 2345060..2346091 | - | 1032 | ANP65424 | UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase | - |
| BAU10_10600 | 2346098..2346607 | - | 510 | ANP66491 | molecular chaperone | - |
| BAU10_10605 | 2346621..2349035 | - | 2415 | ANP65425 | outer membrane protein assembly factor BamA | - |
| BAU10_10610 | 2349080..2350438 | - | 1359 | ANP65426 | RIP metalloprotease RseP | - |
| BAU10_10615 | 2350438..2351643 | - | 1206 | ANP65427 | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | - |
| BAU10_10620 | 2351694..2352536 | - | 843 | ANP65428 | CDP-diglyceride synthetase | - |
| BAU10_10625 | 2352549..2353304 | - | 756 | ANP65429 | di-trans,poly-cis-decaprenylcistransferase | - |
| BAU10_10630 | 2353401..2353958 | - | 558 | ANP65430 | ribosome recycling factor | - |
| BAU10_10635 | 2354059..2354784 | - | 726 | ANP65431 | UMP kinase | - |
| BAU10_10640 | 2354932..2355777 | - | 846 | ANP65432 | translation elongation factor Ts | - |
| BAU10_10645 | 2355910..2356638 | - | 729 | ANP65433 | 30S ribosomal protein S2 | - |
| BAU10_10650 | 2357023..2357901 | + | 879 | ANP65434 | type I methionyl aminopeptidase | - |
| BAU10_10655 | 2358010..2360634 | + | 2625 | ANP65435 | [protein-PII] uridylyltransferase | - |
| BAU10_10660 | 2360777..2361160 | + | 384 | ANP65436 | hypothetical protein | - |
| BAU10_10665 | 2361228..2362010 | - | 783 | ANP65437 | tRNA pseudouridine(65) synthase TruC | - |
| BAU10_10670 | 2362015..2362329 | - | 315 | ANP66492 | hypothetical protein | - |
| BAU10_10675 | 2362406..2363443 | + | 1038 | ANP65438 | hypothetical protein | - |
| BAU10_10680 | 2363457..2363756 | + | 300 | ANP65439 | hypothetical protein | - |
| BAU10_10685 | 2363716..2364222 | - | 507 | ANP65440 | acetyltransferase | - |
| BAU10_10690 | 2365133..2365426 | + | 294 | ANP65441 | hypothetical protein | - |
| BAU10_10695 | 2365423..2365872 | + | 450 | ANP65442 | hypothetical protein | - |
| BAU10_10700 | 2366226..2367056 | + | 831 | ANP66493 | 3-oxoacyl-ACP synthase | - |
| BAU10_10705 | 2367053..2368039 | + | 987 | ANP65443 | UDP-glucose 4-epimerase | - |
| BAU10_10710 | 2368008..2368871 | + | 864 | ANP65444 | polysaccharide biosynthesis protein GumP | - |
| BAU10_10715 | 2368868..2370127 | + | 1260 | ANP66494 | CoF synthetase | - |
| BAU10_10720 | 2370115..2371227 | + | 1113 | ANP65445 | carboxylate--amine ligase | - |
| BAU10_10725 | 2371224..2372297 | + | 1074 | ANP65446 | hypothetical protein | - |
| BAU10_10730 | 2372404..2373990 | - | 1587 | ANP65447 | aminobenzoyl-glutamate transporter | - |
| BAU10_10735 | 2374349..2374609 | - | 261 | ANP65448 | 50S ribosomal protein L31 | - |
| BAU10_10740 | 2374733..2375428 | + | 696 | ANP65449 | tRNA-Thr(GGU) m(6)t(6)A37 methyltransferase TsaA | - |
| BAU10_10745 | 2375540..2377255 | + | 1716 | ANP65450 | proline--tRNA ligase | - |
| BAU10_10750 | 2377432..2378100 | + | 669 | ANP65451 | hypothetical protein | - |
| BAU10_10755 | 2378212..2378418 | + | 207 | ANP65452 | hypothetical protein | - |
| BAU10_10760 | 2378418..2378609 | + | 192 | ANP65453 | hypothetical protein | - |
| BAU10_10765 | 2378714..2380078 | - | 1365 | ANP65454 | transporter | - |
| BAU10_10770 | 2380457..2383003 | + | 2547 | ANP65455 | chitinase | - |
| BAU10_10775 | 2383094..2383837 | - | 744 | ANP65456 | chemotaxis protein | - |
| BAU10_10780 | 2383899..2384378 | - | 480 | ANP65457 | transcription elongation factor | - |
| BAU10_10785 | 2384455..2386380 | + | 1926 | ANP65458 | recombinase RecQ | - |
| BAU10_10790 | 2386377..2387441 | - | 1065 | ANP66495 | DNA polymerase IV | - |
| BAU10_10795 | 2387518..2387751 | - | 234 | ANP65459 | hypothetical protein | - |
| BAU10_10800 | 2387939..2389156 | + | 1218 | ANP65460 | diguanylate cyclase response regulator | - |
| BAU10_10805 | 2389315..2389548 | - | 234 | ANP65461 | hypothetical protein | - |
| BAU10_10810 | 2389571..2390575 | - | 1005 | ANP65462 | FAD:protein FMN transferase ApbE | - |
| BAU10_10815 | 2390733..2391956 | - | 1224 | ANP65463 | NADH:ubiquinone reductase (Na(+)-transporting) subunit F | - |
| BAU10_10820 | 2391990..2392586 | - | 597 | ANP65464 | NADH:ubiquinone reductase (Na(+)-transporting) subunit E | - |
| BAU10_10825 | 2392594..2393226 | - | 633 | ANP65465 | NADH:ubiquinone reductase (Na(+)-transporting) subunit D | - |
| BAU10_10830 | 2393226..2393996 | - | 771 | ANP65466 | Na(+)-translocating NADH-quinone reductase subunit C | - |
| BAU10_10835 | 2393986..2395230 | - | 1245 | ANP65467 | NADH:ubiquinone reductase (Na(+)-transporting) subunit B | - |
| BAU10_10840 | 2395236..2396576 | - | 1341 | ANP66496 | NADH:ubiquinone reductase (Na(+)-transporting) subunit A | - |
| BAU10_10845 | 2397050..2397358 | - | 309 | ANP65468 | transcriptional regulator BolA | - |
| BAU10_10850 | 2397602..2398756 | + | 1155 | ANP65469 | 50S rRNA methyltransferase | - |
| BAU10_10855 | 2398789..2399358 | + | 570 | ANP65470 | hypothetical protein | - |
| BAU10_10860 | 2399358..2399903 | + | 546 | ANP65471 | peptidyl-prolyl cis-trans isomerase | - |
| BAU10_10865 | 2399975..2401327 | + | 1353 | ANP65472 | MFS transporter | - |
| BAU10_10870 | 2401391..2402050 | - | 660 | ANP65473 | transcriptional regulator | - |
| BAU10_10875 | 2402047..2402631 | - | 585 | ANP65474 | RNA polymerase subunit sigma | - |
| BAU10_10880 | 2402676..2404034 | - | 1359 | ANP65475 | 2-dehydropantoate 2-reductase | - |
| BAU10_10885 | 2404031..2404627 | - | 597 | ANP65476 | ATP-dependent protease | - |
| BAU10_10890 | 2404951..2405748 | + | 798 | ANP65477 | hypothetical protein | - |
| BAU10_10895 | 2405857..2406747 | + | 891 | ANP65478 | 2-dehydropantoate 2-reductase | - |
| BAU10_10900 | 2406744..2407343 | + | 600 | ANP65479 | oxidative-stress-resistance chaperone | - |
| BAU10_10905 | 2407343..2408554 | + | 1212 | ANP65480 | cysteine sulfinate desulfinase | - |
| BAU10_10910 | 2408588..2409508 | - | 921 | ANP65481 | diguanylate cyclase | - |
| BAU10_10915 | 2409818..2410246 | - | 429 | ANP65482 | cysteine desulfurase, sulfur acceptor subunit CsdE | - |
| BAU10_10920 | 2410266..2411075 | - | 810 | ANP65483 | tRNA threonylcarbamoyladenosine dehydratase | - |
| BAU10_10925 | 2411175..2412278 | - | 1104 | ANP65484 | murein transglycosylase A | - |
| BAU10_10930 | 2412552..2413007 | - | 456 | ANP65485 | N-acetylglutamate synthase | - |
| BAU10_10935 | 2413170..2414507 | + | 1338 | ANP66497 | amino-acid N-acetyltransferase | - |
| BAU10_10940 | 2415161..2415532 | - | 372 | ANP65486 | hypothetical protein | - |
| BAU10_10945 | 2415520..2416530 | - | 1011 | ANP65487 | peptidoglycan-binding protein LysM | - |
| BAU10_10950 | 2416696..2418843 | - | 2148 | ANP65488 | exodeoxyribonuclease V subunit alpha | - |
| BAU10_10955 | 2418843..2422517 | - | 3675 | ANP66498 | exodeoxyribonuclease V subunit beta | - |
| BAU10_10960 | 2422625..2422810 | - | 186 | ANP65489 | hypothetical protein | - |
| BAU10_10965 | 2422845..2426336 | - | 3492 | ANP65490 | exodeoxyribonuclease V subunit gamma | - |
| BAU10_10970 | 2426589..2428412 | + | 1824 | ANP65491 | hydrolase | - |
| BAU10_10975 | 2428488..2429462 | - | 975 | ANP65492 | C4-dicarboxylate ABC transporter | - |
| BAU10_10980 | 2429572..2430498 | + | 927 | ANP65493 | XRE family transcriptional regulator | - |
| BAU10_10985 | 2430627..2430926 | + | 300 | ANP65494 | multidrug DMT transporter permease | - |
| BAU10_10990 | 2431418..2432110 | + | 693 | ANP65495 | hypothetical protein | - |
| BAU10_10995 | 2432134..2433117 | + | 984 | ANP65496 | GGGtGRT protein | - |
| BAU10_11000 | 2433443..2434474 | + | 1032 | ANP65497 | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | - |
| BAU10_11005 | 2434556..2435611 | - | 1056 | ANP65498 | glycerophosphodiester phosphodiesterase | - |
| BAU10_11010 | 2435668..2437032 | - | 1365 | ANP65499 | glycerol-3-phosphate transporter | - |
| BAU10_11015 | 2437929..2438783 | + | 855 | ANP65500 | aquaporin | - |
| BAU10_11020 | 2438843..2440360 | + | 1518 | ANP65501 | glycerol kinase | - |
| BAU10_11025 | 2440520..2441311 | + | 792 | ANP65502 | transcriptional regulator | - |
| BAU10_11030 | 2441489..2443048 | + | 1560 | ANP65503 | glycerol-3-phosphate dehydrogenase | - |
| BAU10_11035 | 2443325..2444278 | + | 954 | ANP65504 | magnesium transporter | - |
| BAU10_11040 | 2444352..2445536 | - | 1185 | ANP65505 | short-chain dehydrogenase | - |
| BAU10_11045 | 2445719..2446270 | + | 552 | ANP65506 | hypothetical protein | - |
| BAU10_11050 | 2446309..2446896 | - | 588 | ANP65507 | histidine kinase | - |
| BAU10_11055 | 2447317..2448303 | + | 987 | ANP65508 | transcriptional regulator EbgR | - |
| BAU10_11060 | 2448474..2449391 | - | 918 | ANP66499 | LysR family transcriptional regulator | - |
| BAU10_11065 | 2449504..2450643 | + | 1140 | ANP65509 | chorismate-binding protein | - |
| BAU10_11070 | 2450767..2451411 | - | 645 | ANP65510 | helix-turn-helix transcriptional regulator | - |
| BAU10_11075 | 2451980..2452909 | + | 930 | ANP65511 | dentin sialophosphoprotein | - |
| BAU10_11080 | 2452930..2453253 | + | 324 | ANP65512 | carbon storage regulator | - |
| BAU10_11085 | 2453259..2453936 | + | 678 | ANP65513 | bacteriocin resistance protein | - |
| BAU10_11090 | 2453991..2454557 | + | 567 | ANP65514 | hypothetical protein | - |
| BAU10_11095 | 2454782..2455777 | - | 996 | ANP65515 | hypothetical protein | - |
| BAU10_11100 | 2456254..2457945 | + | 1692 | ANP66500 | AMP-dependent synthetase | - |
| BAU10_11105 | 2458321..2459268 | + | 948 | ANP65516 | transcriptional regulator | - |
| BAU10_11110 | 2459375..2461465 | - | 2091 | ANP65517 | translation elongation factor G | - |
| BAU10_11115 | 2461681..2463060 | - | 1380 | ANP65518 | DNA repair protein RadA | - |
| BAU10_11120 | 2463197..2465545 | + | 2349 | ANP65519 | pilus assembly protein PilZ | - |
| BAU10_11125 | 2465609..2466589 | - | 981 | ANP65520 | phosphoserine phosphatase SerB | - |
| BAU10_11130 | 2466681..2467286 | + | 606 | ANP65521 | SMP protein | - |
| BAU10_11135 | 2467382..2468101 | - | 720 | ANP65522 | purine-nucleoside phosphorylase | - |
| BAU10_11140 | 2468245..2469465 | - | 1221 | ANP65523 | phosphopentomutase | - |
| BAU10_11145 | 2469534..2470862 | - | 1329 | ANP65524 | thymidine phosphorylase | - |
| BAU10_11150 | 2470960..2471736 | - | 777 | ANP65525 | deoxyribose-phosphate aldolase | - |
| BAU10_11155 | 2472320..2473603 | - | 1284 | ANP65526 | nucleoside transporter NupC | - |
| BAU10_11160 | 2473808..2474581 | - | 774 | ANP65527 | deoxyribonuclease | - |
| BAU10_11165 | 2474568..2474765 | - | 198 | ANP65528 | hypothetical protein | - |
| BAU10_11170 | 2474860..2476449 | - | 1590 | ANP65529 | peptide chain release factor 3 | - |
| BAU10_11175 | 2476706..2477353 | - | 648 | ANP65530 | transcriptional regulator | - |
| BAU10_11180 | 2477471..2477722 | + | 252 | ANP65531 | Cro/Cl family transcriptional regulator | - |
| BAU10_11185 | 2477778..2478647 | + | 870 | ANP65532 | hypothetical protein | - |
| BAU10_11190 | 2478634..2479296 | + | 663 | ANP65533 | hypothetical protein | - |
| BAU10_11195 | 2479283..2479831 | + | 549 | ANP65534 | transglycosylase | - |
| BAU10_11200 | 2479828..2480127 | + | 300 | ANP65535 | transcriptional regulator | - |
| BAU10_11205 | 2480124..2480657 | + | 534 | ANP65536 | transcriptional regulator | - |
| BAU10_11210 | 2480712..2481143 | + | 432 | ANP65537 | hypothetical protein | - |
| BAU10_11215 | 2481176..2484745 | - | 3570 | ANP65538 | conjugal transfer protein TraG | - |
| BAU10_11220 | 2484749..2486137 | - | 1389 | ANP65539 | conjugal transfer protein TraH | - |
| BAU10_11225 | 2486140..2487084 | - | 945 | ANP65540 | pilus assembly protein | - |
| BAU10_11230 | 2487614..2488204 | + | 591 | ANP65541 | hypothetical protein | - |
| BAU10_11235 | 2488380..2489369 | + | 990 | ANP65542 | hypothetical protein | - |
| BAU10_11240 | 2489369..2492410 | + | 3042 | ANP65543 | hypothetical protein | - |
| BAU10_11245 | 2492411..2492887 | + | 477 | ANP65544 | hypothetical protein | - |
| BAU10_11250 | 2493132..2494280 | + | 1149 | ANP65545 | diguanylate cyclase | - |
| BAU10_11255 | 2494449..2495156 | - | 708 | ANP65546 | hypothetical protein | - |
| BAU10_11260 | 2495246..2496319 | - | 1074 | ANP65547 | DNA primase | - |
| BAU10_11265 | 2496411..2496752 | - | 342 | ANP65548 | plasmid-related protein | - |
| BAU10_11270 | 2496752..2497249 | - | 498 | ANP65549 | DNA repair protein RadC | - |
| BAU10_11275 | 2497332..2498987 | - | 1656 | ANP65550 | hypothetical protein | - |
| BAU10_11280 | 2499057..2499497 | - | 441 | ANP65551 | hypothetical protein | - |
| BAU10_11285 | 2499559..2500512 | - | 954 | ANP65552 | cobalamin biosynthesis protein | - |
| BAU10_11290 | 2500611..2501378 | - | 768 | ANP65553 | hypothetical protein | - |
| BAU10_11295 | 2501378..2502337 | - | 960 | ANP65554 | ATPase | - |
| BAU10_11300 | 2502547..2503563 | - | 1017 | ANP65555 | endonuclease | - |
| BAU10_11305 | 2503849..2504667 | - | 819 | ANP65556 | phage recombination protein Bet | - |
| BAU10_11310 | 2504747..2505166 | - | 420 | ANP65557 | single-stranded DNA-binding protein | - |
| BAU10_11315 | 2505182..2505508 | - | 327 | ANP65558 | plasmid-related protein | - |
| BAU10_11320 | 2505876..2506478 | + | 603 | ANP65559 | hypothetical protein | - |
| BAU10_11325 | 2506569..2507231 | - | 663 | ANP65560 | hypothetical protein | - |
| BAU10_11330 | 2507262..2507585 | - | 324 | ANP66501 | hypothetical protein | - |
| BAU10_11335 | 2507644..2508063 | - | 420 | ANP66502 | S26 family signal peptidase | - |
| BAU10_11340 | 2508140..2508487 | - | 348 | ANP65561 | plasmid-related protein | - |
| BAU10_11345 | 2508617..2509720 | + | 1104 | ANP65562 | transposase | - |
| BAU10_11350 | 2509783..2512194 | - | 2412 | ANP65563 | type-IV secretion system protein TraC | virb4 |
| BAU10_11355 | 2512194..2512886 | - | 693 | ANP65564 | disulfide isomerase | trbB |
| BAU10_11360 | 2513018..2513956 | - | 939 | ANP65565 | hypothetical protein | - |
| BAU10_11365 | 2513949..2514740 | - | 792 | ANP66503 | hypothetical protein | - |
| BAU10_11370 | 2514960..2515346 | - | 387 | ANP65566 | hypothetical protein | - |
| BAU10_11375 | 2515343..2515993 | - | 651 | ANP65567 | conjugal transfer protein TraV | traV |
| BAU10_11380 | 2515990..2517279 | - | 1290 | ANP65568 | conjugal transfer protein TraB | traB |
| BAU10_11385 | 2517286..2518179 | - | 894 | ANP66504 | hypothetical protein | traK |
| BAU10_11390 | 2518163..2518789 | - | 627 | ANP65569 | pilus assembly protein | traE |
| BAU10_11395 | 2518786..2519067 | - | 282 | ANP65570 | type IV conjugative transfer system protein TraL | traL |
| BAU10_11400 | 2519222..2520427 | - | 1206 | ANP65571 | transposase | - |
| BAU10_11405 | 2520503..2521609 | - | 1107 | ANP65572 | zinc peptidase | - |
| BAU10_11410 | 2521602..2522186 | - | 585 | ANP65573 | hypothetical protein | - |
| BAU10_11415 | 2522312..2522947 | - | 636 | ANP65574 | conjugal transfer protein | tfc7 |
| BAU10_11420 | 2522934..2523494 | - | 561 | ANP65575 | conjugative transfer protein | - |
| BAU10_11425 | 2523504..2525324 | - | 1821 | ANP65576 | conjugal transfer protein | virb4 |
| BAU10_11430 | 2525373..2527523 | - | 2151 | ANP65577 | phosphohydrolase | - |
| BAU10_11435 | 2527680..2528861 | - | 1182 | ANP65578 | hypothetical protein | - |
Host bacterium
| ID | 355 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEValZJT1 | GenBank | CP016224 |
| Element size | 3535128 bp | Coordinate of oriT [Strand] | 84579..84877 [-] |
| Host bacterium | Vibrio alginolyticus strain ZJ-T | Coordinate of element | 2476706..2564476 |
Cargo genes
| Drug resistance gene | sul2, aph(3'')-Ib, aph(6)-Id, dfrA31, qnrVC5 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |