Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200396 |
| Name | oriT_ICESsuYS388 |
| Organism | Streptococcus suis sequence |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | MK211824 (37889..37923 [-], 35 nt) |
| oriT length | 35 nt |
| IRs (inverted repeats) | 5..11, 15..21 (ACTTTGC..GCAAAGT) |
| Location of nic site | 29..30 |
| Conserved sequence flanking the nic site |
GTGTGTTATA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 35 nt
>oriT_ICESsuYS388
CACGACTTTGCGAAGCAAAGTGTAGTGTGTTATAC
CACGACTTTGCGAAGCAAAGTGTAGTGTGTTATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file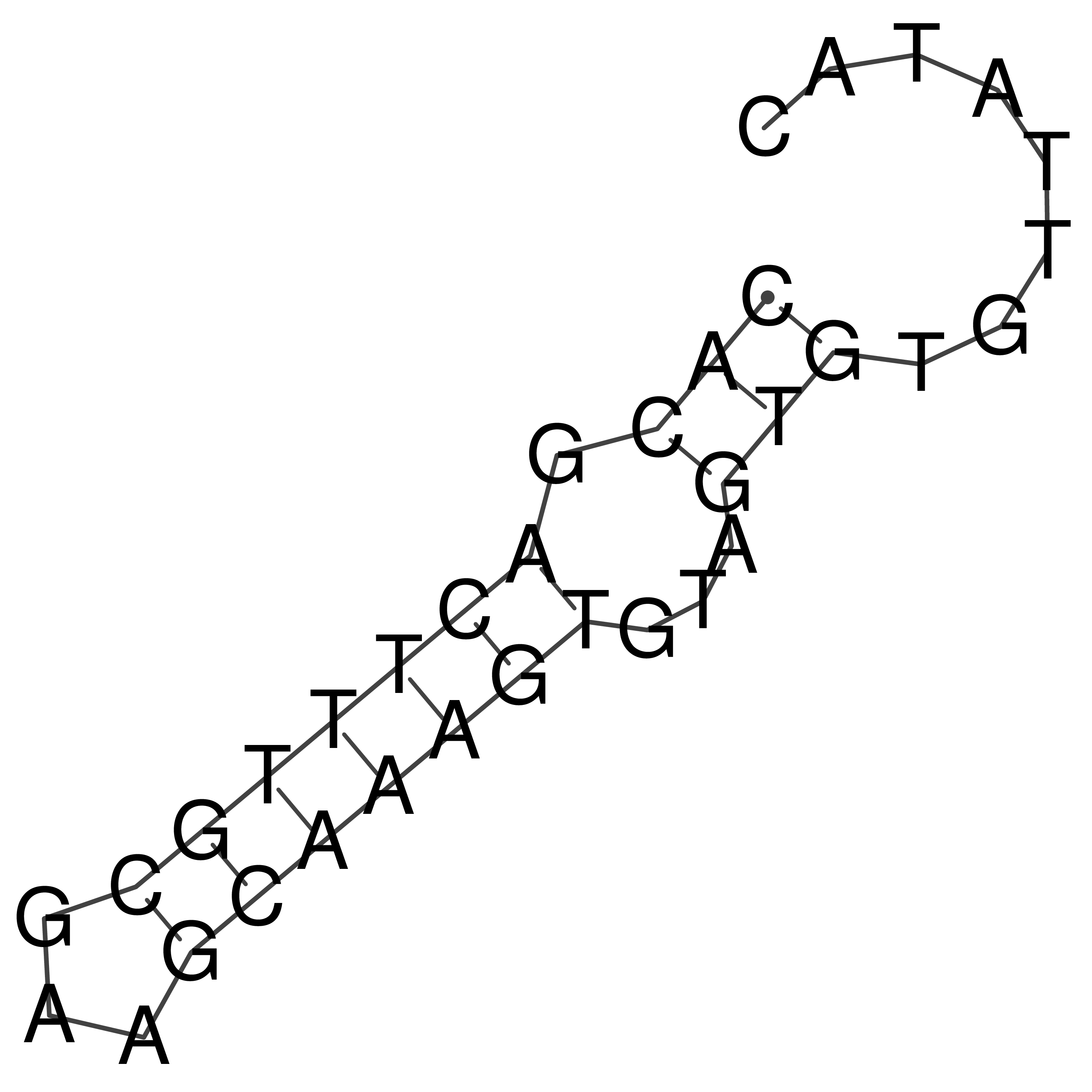
Relaxase
| ID | 14422 | GenBank | QID26664 |
| Name | Relaxase_YS388-GM000016_ICESsuYS388 |
UniProt ID | _ |
| Length | 627 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 627 a.a. Molecular weight: 74663.90 Da Isoelectric Point: 7.1885
>QID26664.1 relaxase [Streptococcus suis]
MVVTKHFAVHGNKYRKSLIKYILNPAKTDQLKLVSDFGMSNYLDFPNYEEIVEMYQANFVNNDRLYDSRN
DRQQIKQNKIHAHHLIQSFSPEDNLTPEEINRIGYETIKELTSGNFRFIVTTHTDRLHVHNHILINSVDL
NSHKKLKWDYAQERNLRMISDRLSKEAGAKIITPNRYSHEKFVTYRKSNHKFELKQRLYFLMENSKDFDD
FLSKAEALNVQIDFSRKYARFLMTDMPMKRVIRGKQLDKRQPYTEEYFREHFAKRAIEQCLDFLLPRVRD
LSQLLEFARELNLAMSLKQKNVAFTLTKNSQSITVNNQKVSSKNLYDVQFFETYFEKRGEVPDIDQSQLI
SDFEGYCEEQDKEKLAYEDLQEAYQAFKEKRDQVQEFEVVLADHQIDKLVKDGLFIRMNYGVKKDGLVFI
PNRHLDIKETESGKHYHAFIRETAQFFIYNKEASELNRYMRGRELIRQLTNDSQTISKRKRPTIDTLKKK
IEEINLLIELGTENKSYQDIKDEIIKDIAQLDLTITEIQEHIDHLNKVAEVLLNLDNNDIENRRLAKYDY
AKMNLTAAIKLEQVEKEIEIYQIKLDKSIDDYEYLIRRLEKFVKILNEIDSSKDELKFEHSEKNVKE
MVVTKHFAVHGNKYRKSLIKYILNPAKTDQLKLVSDFGMSNYLDFPNYEEIVEMYQANFVNNDRLYDSRN
DRQQIKQNKIHAHHLIQSFSPEDNLTPEEINRIGYETIKELTSGNFRFIVTTHTDRLHVHNHILINSVDL
NSHKKLKWDYAQERNLRMISDRLSKEAGAKIITPNRYSHEKFVTYRKSNHKFELKQRLYFLMENSKDFDD
FLSKAEALNVQIDFSRKYARFLMTDMPMKRVIRGKQLDKRQPYTEEYFREHFAKRAIEQCLDFLLPRVRD
LSQLLEFARELNLAMSLKQKNVAFTLTKNSQSITVNNQKVSSKNLYDVQFFETYFEKRGEVPDIDQSQLI
SDFEGYCEEQDKEKLAYEDLQEAYQAFKEKRDQVQEFEVVLADHQIDKLVKDGLFIRMNYGVKKDGLVFI
PNRHLDIKETESGKHYHAFIRETAQFFIYNKEASELNRYMRGRELIRQLTNDSQTISKRKRPTIDTLKKK
IEEINLLIELGTENKSYQDIKDEIIKDIAQLDLTITEIQEHIDHLNKVAEVLLNLDNNDIENRRLAKYDY
AKMNLTAAIKLEQVEKEIEIYQIKLDKSIDDYEYLIRRLEKFVKILNEIDSSKDELKFEHSEKNVKE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14423 | GenBank | QID26683 |
| Name | Mob_Pre_YS388-GM000037_ICESsuYS388 |
UniProt ID | _ |
| Length | 383 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 383 a.a. Molecular weight: 43958.43 Da Isoelectric Point: 10.1155
>QID26683.1 plasmid recombination protein, Mob family [Streptococcus suis]
MTVTALARGYPKNTVIIDMTVMQGVRVTVAAVGRGDPKTAVTTVTAVTPKDGHPPKERRNPPMPYAILRF
QKRKAGGVAACERHNERKKEAYKSNPDIDMERSKNNYHLIAPPKYTYKKEINRMVAEAGCRTRKDSVMMV
ETLITASPEFMNQLPPEEQKAYFQTALDFISERVGKQNILSAVVHMDERTPHMHLCFVPITPDNKLSAKA
ILGNQKSLSEWQTAYHERMSSRWNQLERGQSSMETKRKHVPTWLYKLGGRLDKQYEEIVSALSDINAFNA
GKKRDKALDLLSAWLPDVEKFSKEIGKQQAYIDSLKERIGQESDYAGRMRDEKYEQELKVQKANQKIFEL
QRTNEQMGRLLSKIPPEVLEELQKNHRSRAKER
MTVTALARGYPKNTVIIDMTVMQGVRVTVAAVGRGDPKTAVTTVTAVTPKDGHPPKERRNPPMPYAILRF
QKRKAGGVAACERHNERKKEAYKSNPDIDMERSKNNYHLIAPPKYTYKKEINRMVAEAGCRTRKDSVMMV
ETLITASPEFMNQLPPEEQKAYFQTALDFISERVGKQNILSAVVHMDERTPHMHLCFVPITPDNKLSAKA
ILGNQKSLSEWQTAYHERMSSRWNQLERGQSSMETKRKHVPTWLYKLGGRLDKQYEEIVSALSDINAFNA
GKKRDKALDLLSAWLPDVEKFSKEIGKQQAYIDSLKERIGQESDYAGRMRDEKYEQELKVQKANQKIFEL
QRTNEQMGRLLSKIPPEVLEELQKNHRSRAKER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16944 | GenBank | QID26706 |
| Name | t4cp2_YS388-GM000060_ICESsuYS388 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 69106.73 Da Isoelectric Point: 9.2044
>QID26706.1 conjugal transfer protein TraG [Streptococcus suis]
MVSGKKVFIFGILGLALGYFCHRLVLLYDSLPNQPPLERLAYLLGEGQNQVLNPLWNGNFTGKSVLGFCF
GFVTMGLVYLYVSTGQKVYREGAEYGSARFGNNRERKAFLSKNPFNDTILSRNVRLTLLERKAPQFDRNK
NLVVIGGSGAGKTFRFVKPNLIQLNCSNIVVDPKDHLAEKTGKLFLEHGYQVKVLDLVNMTNSDGFNPFR
YVETENDLNRMLTVYFNNTKGNGSRSDPFWDEASMTLVRAISSYLVDFYNPPGSTKEEADSRRKRGRYPS
FSEIGKLIKLLSKGENQDKSVLEVMFETYAKTYGTENFTMRNWADFQNYKDKTLDSVIAVTTAKFALFNI
QSVIDLTKRDTLDLKTWGTQKTMVYLVIPDNDTTFRFLSALFFSTVFSTLTRQADVDFKGQLPIHVRSYL
DEFANVGEIPDFAEQTSTVRSRNMSLVPILQNIAQLQGLYKEKEAWKTILGNCDSLLYLGGNDEETFKFM
SGLLGKQTIDVRSTSRSYGQTGSGSTSHQKIARDLMTPDEVGNMKRDECLVRIAGVPVFKEKKYFPLKHK
NWKYLADKESDERWWHYHIDPLKTEDVPFEPSDHKVRDLSKESTLH
MVSGKKVFIFGILGLALGYFCHRLVLLYDSLPNQPPLERLAYLLGEGQNQVLNPLWNGNFTGKSVLGFCF
GFVTMGLVYLYVSTGQKVYREGAEYGSARFGNNRERKAFLSKNPFNDTILSRNVRLTLLERKAPQFDRNK
NLVVIGGSGAGKTFRFVKPNLIQLNCSNIVVDPKDHLAEKTGKLFLEHGYQVKVLDLVNMTNSDGFNPFR
YVETENDLNRMLTVYFNNTKGNGSRSDPFWDEASMTLVRAISSYLVDFYNPPGSTKEEADSRRKRGRYPS
FSEIGKLIKLLSKGENQDKSVLEVMFETYAKTYGTENFTMRNWADFQNYKDKTLDSVIAVTTAKFALFNI
QSVIDLTKRDTLDLKTWGTQKTMVYLVIPDNDTTFRFLSALFFSTVFSTLTRQADVDFKGQLPIHVRSYL
DEFANVGEIPDFAEQTSTVRSRNMSLVPILQNIAQLQGLYKEKEAWKTILGNCDSLLYLGGNDEETFKFM
SGLLGKQTIDVRSTSRSYGQTGSGSTSHQKIARDLMTPDEVGNMKRDECLVRIAGVPVFKEKKYFPLKHK
NWKYLADKESDERWWHYHIDPLKTEDVPFEPSDHKVRDLSKESTLH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 41169..66897
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| YS388-GM000043 | 36609..37475 | + | 867 | QID26689 | aminoglycoside 6-adenylyltransferase | - |
| YS388-GM000044 | 38511..40430 | - | 1920 | QID26690 | TetO | - |
| YS388-GM000045 | 40814..41188 | - | 375 | QID26691 | TnpV protein | - |
| YS388-GM000046 | 41169..41858 | - | 690 | QID26692 | hypothetical protein | prgL |
| YS388-GM000047 | 41954..42244 | - | 291 | QID26693 | hypothetical protein | gbs1350 |
| YS388-GM000048 | 42258..42557 | - | 300 | QID26694 | hypothetical protein | - |
| YS388-GM000049 | 42628..49446 | - | 6819 | QID26695 | SNF2 family protein | - |
| YS388-GM000050 | 49496..50047 | - | 552 | QID26696 | hypothetical protein | gbs1354 |
| YS388-GM000051 | 50031..50222 | - | 192 | QID26697 | calcium-binding protein | - |
| YS388-GM000052 | 50223..55127 | - | 4905 | QID26698 | glucan-binding protein | prgB |
| YS388-GM000053 | 55124..55228 | - | 105 | QID26699 | hypothetical protein | - |
| YS388-GM000054 | 55370..55963 | - | 594 | QID26700 | hypothetical protein | - |
| YS388-GM000055 | 56208..59012 | - | 2805 | QID26701 | amidase | prgK |
| YS388-GM000056 | 59009..61339 | - | 2331 | QID26702 | VirB4 | virb4 |
| YS388-GM000057 | 61317..61670 | - | 354 | QID26703 | PrgI family protein | prgIc |
| YS388-GM000058 | 61732..62586 | - | 855 | QID26704 | conjugal transfer protein TrbL | prgHb |
| YS388-GM000059 | 62603..62845 | - | 243 | QID26705 | hypothetical protein | prgF |
| YS388-GM000060 | 62863..64683 | - | 1821 | QID26706 | conjugal transfer protein TraG | virb4 |
| YS388-GM000061 | 64683..65171 | - | 489 | QID26707 | hypothetical protein | gbs1365 |
| YS388-GM000062 | 65249..65830 | - | 582 | QID26708 | abortive infection protein | - |
| YS388-GM000063 | 65833..66066 | - | 234 | QID26709 | Uncharacterized protein | - |
| YS388-GM000064 | 66076..66459 | - | 384 | QID26710 | arsenate reductase | - |
| YS388-GM000065 | 66469..66897 | - | 429 | QID26711 | hypothetical protein | gbs1369 |
| YS388-GM000066 | 66881..68245 | - | 1365 | QID26712 | DNA (cytosine-5-)-methyltransferase | - |
| YS388-GM000067 | 68394..69218 | - | 825 | QID26713 | replication initiator protein | - |
| YS388-GM000068 | 69215..69568 | - | 354 | QID26714 | hypothetical protein | - |
| YS388-GM000069 | 69648..70013 | - | 366 | QID26715 | LSU ribosomal protein L7/L12 (P1/P2) rplL | - |
Host bacterium
| ID | 352 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICESsuYS388 | GenBank | MK211824 |
| Element size | 70013 bp | Coordinate of oriT [Strand] | 37889..37923 [-] |
| Host bacterium | Streptococcus suis sequence | Coordinate of element | 1..70013 |
Cargo genes
| Drug resistance gene | erm(B), ant(6)-Ia, tet(O) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |