Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200391 |
| Name | oriT_ICESpsSUT-286-2 |
| Organism | Streptococcus parasuis SUT-286 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | AP024276 (39110..39242 [-], 133 nt) |
| oriT length | 133 nt |
| IRs (inverted repeats) | 65..70, 83..88 (AAATCC..GGATTT) 6..12, 23..29 (ACCCCCC..GGGGGGT) |
| Location of nic site | 75..76 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 133 nt
>oriT_ICESpsSUT-286-2
ACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
ACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file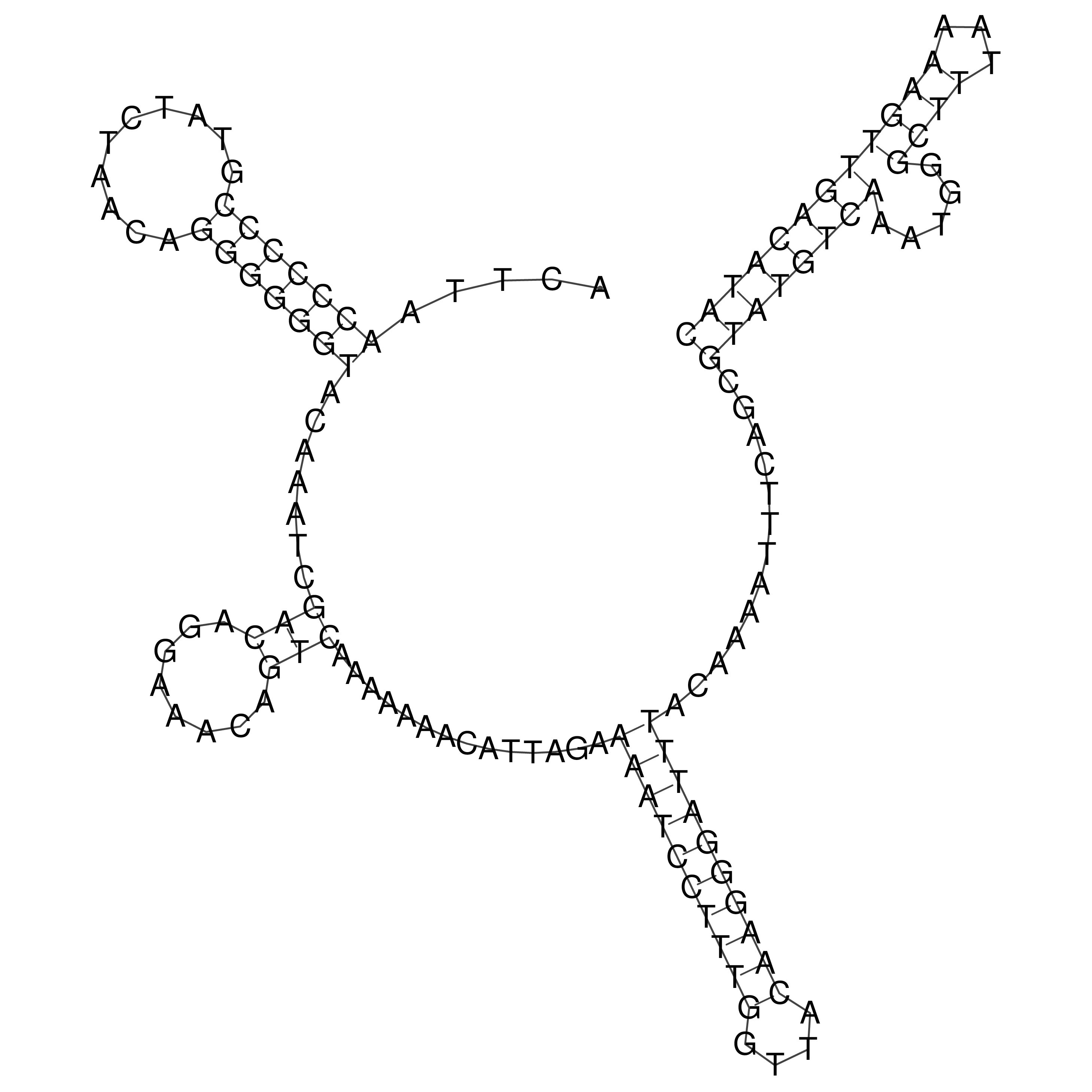
Relaxase
| ID | 14416 | GenBank | BCP60433 |
| Name | Rep_trans_SUT286_17590_ICESpsSUT-286-2 |
UniProt ID | _ |
| Length | 371 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 371 a.a. Molecular weight: 43729.94 Da Isoelectric Point: 6.6982
>BCP60433.1 Cro/Cl family transcriptional regulator [Streptococcus parasuis]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVAGGVM
KRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRSGELVRKEEKECMGNTLYIGSLQSEVYFCIY
EKDYEQYKKNDIPIEDAEVKKRFEIRLKNERAYYAVRDLLVYDNPEHTAFKIINRYIRFVDKDDSKPRSD
WKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAPTLKVAIKLDEINQTQVVKDILDHAKLTD
RHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVAGGVM
KRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRSGELVRKEEKECMGNTLYIGSLQSEVYFCIY
EKDYEQYKKNDIPIEDAEVKKRFEIRLKNERAYYAVRDLLVYDNPEHTAFKIINRYIRFVDKDDSKPRSD
WKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAPTLKVAIKLDEINQTQVVKDILDHAKLTD
RHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7886 | GenBank | BCP60420 |
| Name | BCP60420_ICESpsSUT-286-2 |
UniProt ID | _ |
| Length | 405 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 405 a.a. Molecular weight: 47021.92 Da Isoelectric Point: 9.7978
>BCP60420.1 integrase family protein [Streptococcus parasuis]
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16938 | GenBank | BCP60434 |
| Name | ftsK_3_SUT286_17600_ICESpsSUT-286-2 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>BCP60434.1 cell division protein FtsK [Streptococcus parasuis]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLIYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLIYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1789239..1800559
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SUT286_17470 | 1784426..1784629 | - | 204 | BCP60421 | transposase | - |
| SUT286_17480 | 1785090..1785320 | - | 231 | BCP60422 | hypothetical protein | - |
| SUT286_17490 | 1785317..1785739 | - | 423 | BCP60423 | hypothetical protein | - |
| SUT286_17500 | 1786244..1786597 | + | 354 | BCP60424 | transcriptional regulator | - |
| SUT286_17510 | 1786943..1788862 | - | 1920 | BCP60425 | tetracycline resistance protein TetM | - |
| SUT286_17520 | 1789239..1790171 | - | 933 | BCP60426 | transposase | orf13 |
| SUT286_17530 | 1790168..1791169 | - | 1002 | BCP60427 | NlpC/P60 family hydrolase | orf14 |
| SUT286_17540 | 1791166..1793343 | - | 2178 | BCP60428 | transposase | orf15 |
| SUT286_17550 | 1793346..1795793 | - | 2448 | BCP60429 | ATP/GTP-binding protein | virb4 |
| SUT286_17560 | 1795777..1796169 | - | 393 | BCP60430 | membrane protein | orf17a |
| SUT286_17570 | 1796258..1796755 | - | 498 | BCP60431 | antirestriction protein ArdA | - |
| SUT286_17580 | 1796872..1797093 | - | 222 | BCP60432 | conjugal transfer protein | orf19 |
| SUT286_17590 | 1797136..1798251 | - | 1116 | BCP60433 | Cro/Cl family transcriptional regulator | - |
| SUT286_17600 | 1798429..1799814 | - | 1386 | BCP60434 | cell division protein FtsK | virb4 |
| SUT286_17610 | 1799843..1800229 | - | 387 | BCP60435 | transposase | orf23 |
| SUT286_17620 | 1800245..1800559 | - | 315 | BCP60436 | transposase | orf23 |
| SUT286_17630 | 1801488..1802756 | - | 1269 | BCP60437 | MATE family efflux transporter | - |
| SUT286_17640 | 1802949..1804352 | - | 1404 | BCP60438 | threonine synthase | - |
| SUT286_17650 | 1804507..1805412 | - | 906 | BCP60439 | oligopeptide ABC transporter permease protein | - |
Host bacterium
| ID | 348 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICESpsSUT-286-2 | GenBank | AP024276 |
| Element size | 2197342 bp | Coordinate of oriT [Strand] | 39110..39242 [-] |
| Host bacterium | Streptococcus parasuis SUT-286 | Coordinate of element | 1759153..1810570 |
Cargo genes
| Drug resistance gene | tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |