Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200387 |
| Name | oriT_ICERanRCAD0179-1 |
| Organism | Riemerella anatipestifer strain RCAD0179 Scaffold4 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | QXQM01000004 (18949..19145 [+], 197 nt) |
| oriT length | 197 nt |
| IRs (inverted repeats) | 10..17, 20..27 (TATGTTCA..TGAACATA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 197 nt
>oriT_ICERanRCAD0179-1
ACGGATTTTTATGTTCACCTGAACATAGCAAGATGTCTTTTCAGCCGCTCAAAATATTTTGAGCAGCCACGAAAAGCACTTGCCCTTGCAGGGGGCGGGCTTTCCCTCCGAAGTCGGGAAGCCTTTCAGAACTGCGGTGCTGTAATCCGAAGCGGCGGCTTATGCCGTCAATCCGCTAAATCCAAAGTAATATGAAA
ACGGATTTTTATGTTCACCTGAACATAGCAAGATGTCTTTTCAGCCGCTCAAAATATTTTGAGCAGCCACGAAAAGCACTTGCCCTTGCAGGGGGCGGGCTTTCCCTCCGAAGTCGGGAAGCCTTTCAGAACTGCGGTGCTGTAATCCGAAGCGGCGGCTTATGCCGTCAATCCGCTAAATCCAAAGTAATATGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file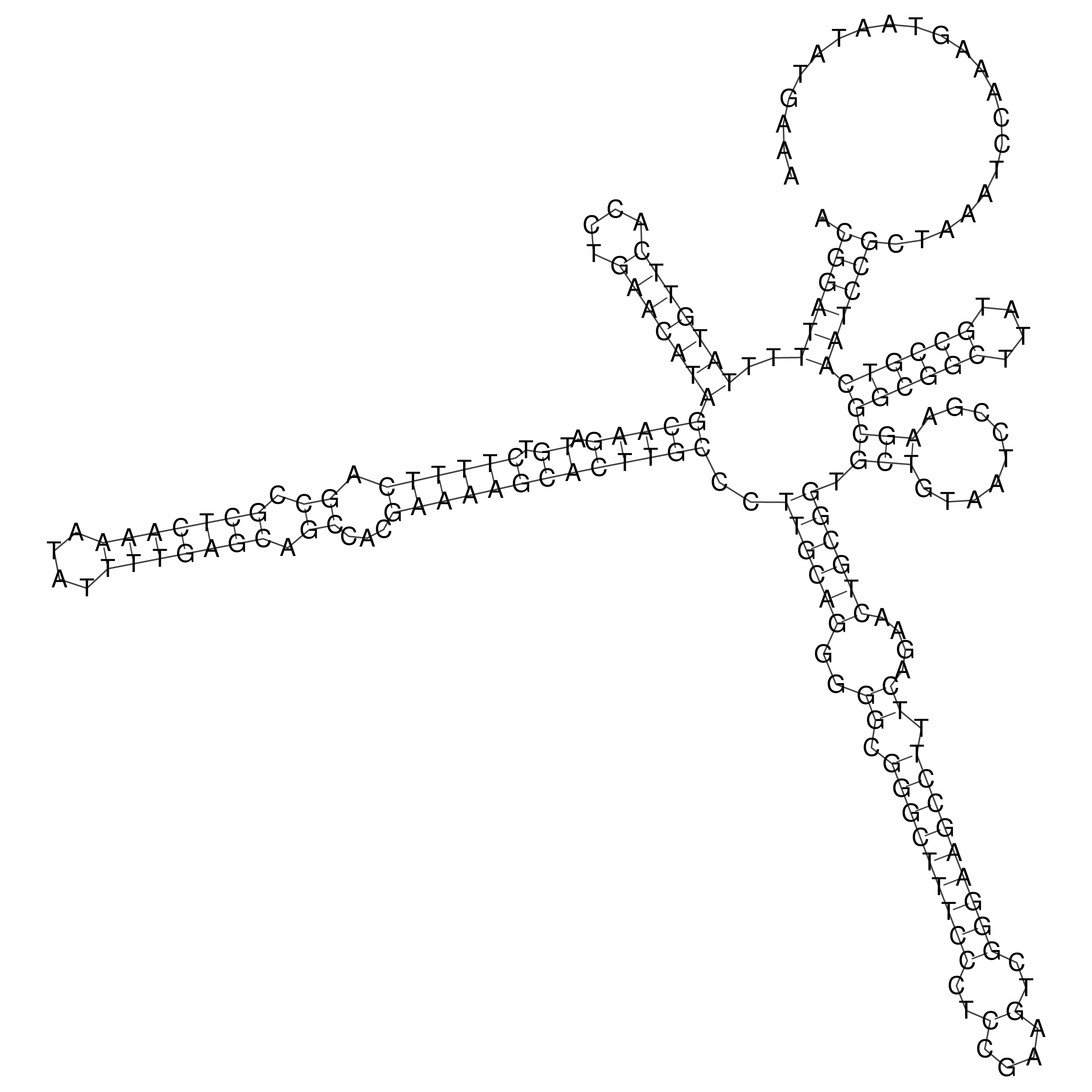
Relaxase
| ID | 14414 | GenBank | MSN90297 |
| Name | Relaxase_D1Y74_04675_ICERanRCAD0179-1 |
UniProt ID | _ |
| Length | 415 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 415 a.a. Molecular weight: 46563.08 Da Isoelectric Point: 9.8931
>MSN90297.1 relaxase [Riemerella anatipestifer]
MVAKISVGSSLYGAIAYNGEKINEAQGRLLTTNRIYNDGSGTVDIGKAMEGFLTFLPPQMKIEKPVVHIS
LNPHPEDVLTDIELQNIAREYLEKLGFGNQPYLVFKHEDIDRHHLHIVTVNVDENGKRLNRDFLYRRSDR
IRRELEQKYGLHPAERKNQRLDNPLRKVAASAGDVKKQVGNTVKALNGQYRFQTMGEYRALLSLYNMTVE
EARGNVRGREYHGLVYSVTDDKGNKVGNPFKSSLFGKSAGYEAVQKKFVRSKSEIKDRKLADMTKRTVLS
VLQGTYDKDKFVSQLKEKGIDTVLRYTEEGRIYGATFIDHRTGCVLNGSRMGKELSANALQEHFTLPYAG
QPPIPLSIPVDAADKAHGQTAYDSEDISGGMGLLTPEGPAVDAEEEAFIRAMKRKKKKKRKGLGM
MVAKISVGSSLYGAIAYNGEKINEAQGRLLTTNRIYNDGSGTVDIGKAMEGFLTFLPPQMKIEKPVVHIS
LNPHPEDVLTDIELQNIAREYLEKLGFGNQPYLVFKHEDIDRHHLHIVTVNVDENGKRLNRDFLYRRSDR
IRRELEQKYGLHPAERKNQRLDNPLRKVAASAGDVKKQVGNTVKALNGQYRFQTMGEYRALLSLYNMTVE
EARGNVRGREYHGLVYSVTDDKGNKVGNPFKSSLFGKSAGYEAVQKKFVRSKSEIKDRKLADMTKRTVLS
VLQGTYDKDKFVSQLKEKGIDTVLRYTEEGRIYGATFIDHRTGCVLNGSRMGKELSANALQEHFTLPYAG
QPPIPLSIPVDAADKAHGQTAYDSEDISGGMGLLTPEGPAVDAEEEAFIRAMKRKKKKKRKGLGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16936 | GenBank | MSN90298 |
| Name | t4cp2_D1Y74_04680_ICERanRCAD0179-1 |
UniProt ID | _ |
| Length | 672 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 672 a.a. Molecular weight: 77086.10 Da Isoelectric Point: 7.1001
>MSN90298.1 conjugal transfer protein TraG [Riemerella anatipestifer]
MSQQEDDLRALAKIMDFLRAVSIILVVMNVYWFCYEAIRLWGVNIGVVDKILLNFDRTAGLFHSILYTKL
FSVLLLALSCLGTKGVKGEKITWGRIWTAFAVGFVLFFLNWWLLPLPLPLEAVTGLYVLTIGTGYVCLLM
GGLWMSRLLKHNLMEDVFNNENESFMQETRLIESEYSVNLPTRFYYRKRWNNGWINVVNPFRASIVLGTP
GSGKSYAVVNNFIKQQIEKGFSQYIYDFKYPDLSTIAYNHLLNHPDGYKVKPKFYVINFDDPRRSHRCNP
IHPDFMEDITDAYESAYTIMLNLNKTWVQKQGDFFVESPIILFASIIWYLKIYQNGKFCTFPHAIEFLNR
RYEDIFPILTSYPELENYLSPFMDAWLGGAAEQLMGQIASAKIPLSRMISPQLYWVMSDSEFTLDINNPE
EPKILCVGNNPDRQNIYGAALGLYNSRIVKLINKKGMLKSSVIIDELPTIYFKGLDNLIATARSNKVAVC
LGFQDFSQLVRDYGDKEAKVVMNTVGNIFSGQVVGETAKTLSERFGKVLQKRQSISINRQDVSTSINTQM
DALIPPSKISGLTQGMFVGSVSDNFNERIEQKIFHCEIVVDAEKVKREESAYKKIPVITNFTDEDGNDRM
KETVQANYRRIKEEVKQIVQEELERIKNDPVLCKLLPDNETV
MSQQEDDLRALAKIMDFLRAVSIILVVMNVYWFCYEAIRLWGVNIGVVDKILLNFDRTAGLFHSILYTKL
FSVLLLALSCLGTKGVKGEKITWGRIWTAFAVGFVLFFLNWWLLPLPLPLEAVTGLYVLTIGTGYVCLLM
GGLWMSRLLKHNLMEDVFNNENESFMQETRLIESEYSVNLPTRFYYRKRWNNGWINVVNPFRASIVLGTP
GSGKSYAVVNNFIKQQIEKGFSQYIYDFKYPDLSTIAYNHLLNHPDGYKVKPKFYVINFDDPRRSHRCNP
IHPDFMEDITDAYESAYTIMLNLNKTWVQKQGDFFVESPIILFASIIWYLKIYQNGKFCTFPHAIEFLNR
RYEDIFPILTSYPELENYLSPFMDAWLGGAAEQLMGQIASAKIPLSRMISPQLYWVMSDSEFTLDINNPE
EPKILCVGNNPDRQNIYGAALGLYNSRIVKLINKKGMLKSSVIIDELPTIYFKGLDNLIATARSNKVAVC
LGFQDFSQLVRDYGDKEAKVVMNTVGNIFSGQVVGETAKTLSERFGKVLQKRQSISINRQDVSTSINTQM
DALIPPSKISGLTQGMFVGSVSDNFNERIEQKIFHCEIVVDAEKVKREESAYKKIPVITNFTDEDGNDRM
KETVQANYRRIKEEVKQIVQEELERIKNDPVLCKLLPDNETV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 60072..70627
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| D1Y74_04520 | 55143..55553 | + | 411 | MSN90268 | hypothetical protein | - |
| D1Y74_04525 | 55565..55809 | - | 245 | Protein_45 | hypothetical protein | - |
| D1Y74_04530 | 55715..55930 | + | 216 | MSN90269 | hypothetical protein | - |
| D1Y74_04535 | 55942..56163 | + | 222 | MSN90270 | hypothetical protein | - |
| D1Y74_04540 | 56177..56593 | + | 417 | MSN90271 | PcfK-like protein | - |
| D1Y74_04545 | 56590..57921 | + | 1332 | MSN90272 | hypothetical protein | - |
| D1Y74_04550 | 57946..58203 | + | 258 | MSN90273 | hypothetical protein | - |
| D1Y74_04555 | 58223..58456 | + | 234 | MSN90274 | DUF3873 domain-containing protein | - |
| D1Y74_04560 | 58453..58698 | + | 246 | MSN90275 | hypothetical protein | - |
| D1Y74_04565 | 58752..59060 | + | 309 | MSN90276 | hypothetical protein | - |
| D1Y74_04570 | 59094..59339 | - | 246 | MSN90277 | hypothetical protein | - |
| D1Y74_04575 | 59548..60075 | - | 528 | MSN90278 | lysozyme | - |
| D1Y74_04580 | 60072..60575 | - | 504 | MSN90279 | DUF3872 domain-containing protein | traQ |
| D1Y74_04585 | 60572..61474 | - | 903 | MSN90280 | DNA primase | traP |
| D1Y74_04590 | 61482..62057 | - | 576 | MSN90281 | conjugal transfer protein TraO | traO |
| D1Y74_04595 | 62060..63046 | - | 987 | MSN90282 | conjugative transposon protein TraN | traN |
| D1Y74_04600 | 63081..64433 | - | 1353 | MSN90283 | conjugative transposon protein TraM | traM |
| D1Y74_04605 | 64414..65352 | - | 939 | Protein_61 | conjugative transposon protein TraK | - |
| D1Y74_04610 | 65384..66388 | - | 1005 | MSN90284 | conjugative transposon protein TraJ | traJ |
| D1Y74_04615 | 66392..67021 | - | 630 | MSN90285 | DUF4141 domain-containing protein | traI |
| D1Y74_04620 | 67045..67425 | - | 381 | MSN90286 | DUF3876 domain-containing protein | traH |
| D1Y74_04625 | 67466..69970 | - | 2505 | MSN90287 | TraG family conjugative transposon ATPase | virb4 |
| D1Y74_04630 | 69967..70299 | - | 333 | MSN90288 | DUF4133 domain-containing protein | traF |
| D1Y74_04635 | 70310..70627 | - | 318 | MSN90289 | DUF4134 domain-containing protein | traE |
| D1Y74_04640 | 70829..71557 | - | 729 | MSN90290 | conjugal transfer protein TraD | - |
| D1Y74_04645 | 71574..71927 | - | 354 | MSN90291 | DUF3408 domain-containing protein | - |
| D1Y74_04650 | 71930..72370 | - | 441 | MSN90292 | DUF3408 domain-containing protein | - |
| D1Y74_04655 | 72373..73422 | - | 1050 | MSN90293 | ParA family protein | - |
| D1Y74_04660 | 73407..73574 | + | 168 | MSN90294 | hypothetical protein | - |
| D1Y74_04665 | 73623..73847 | + | 225 | MSN90295 | hypothetical protein | - |
| D1Y74_04670 | 73844..74272 | + | 429 | MSN90296 | MobA protein | - |
| D1Y74_04675 | 74251..75498 | + | 1248 | MSN90297 | relaxase | - |
Host bacterium
| ID | 344 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICERanRCAD0179-1 | GenBank | QXQM01000004 |
| Element size | 208058 bp | Coordinate of oriT [Strand] | 18949..19145 [+] |
| Host bacterium | Riemerella anatipestifer strain RCAD0179 Scaffold4 | Coordinate of element | 54705..103871 |
Cargo genes
| Drug resistance gene | tet(Q) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9 |