Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200382 |
| Name | oriT_ICEPmiFra1 |
| Organism | Proteus mirabilis sequence |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | MF490434 (4390..4688 [+], 299 nt) |
| oriT length | 299 nt |
| IRs (inverted repeats) | 234..242, 254..262 (AAAGCCAAA..TTTGGCTTT) 45..50, 52..57 (CAAACG..CGTTTG) 7..12, 26..31 (TTTGGC..GCCAAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 299 nt
>oriT_ICEPmiFra1
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file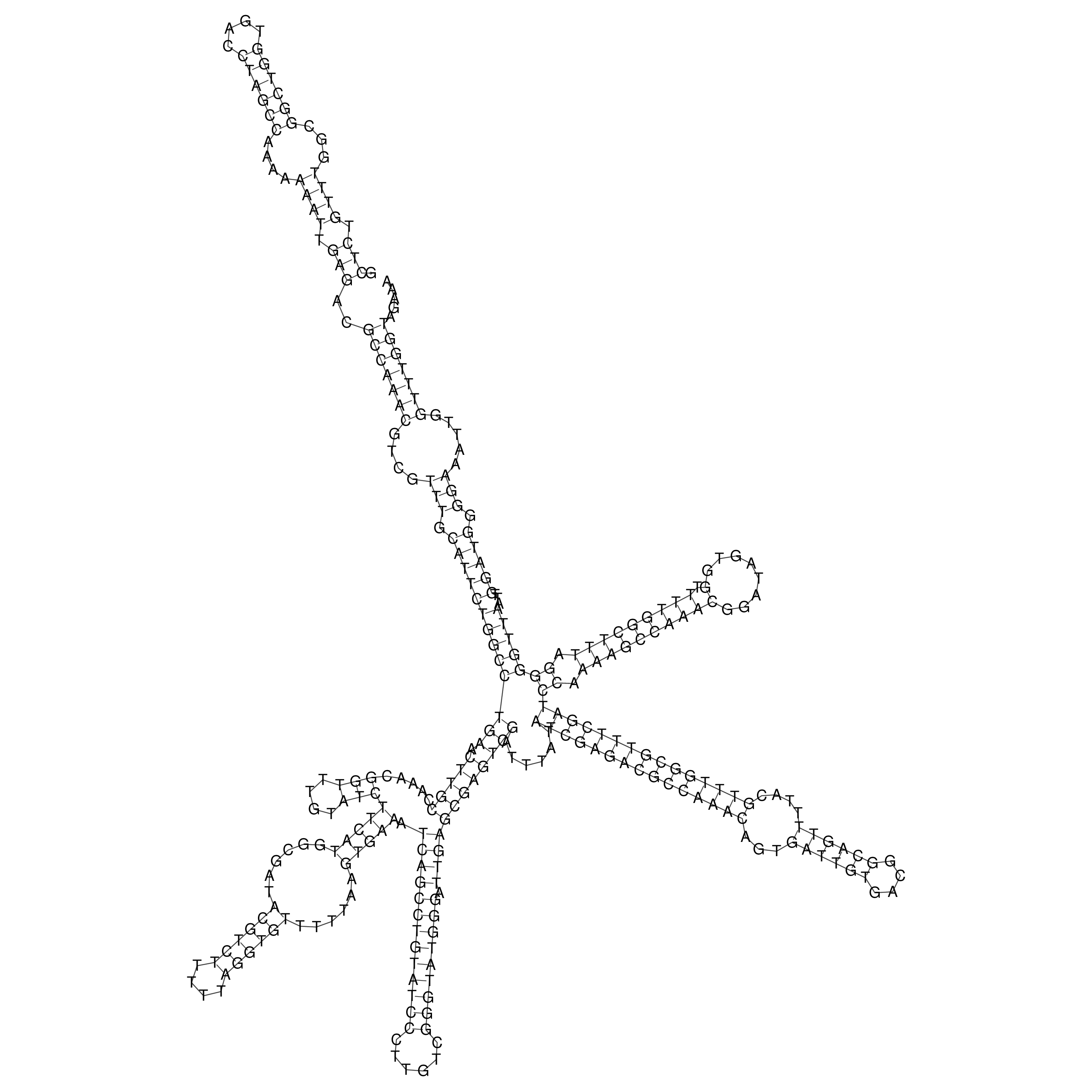
Relaxase
| ID | 14408 | GenBank | AVR61311 |
| Name | traI_-_ICEPmiFra1 |
UniProt ID | _ |
| Length | 716 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 716 a.a. Molecular weight: 79760.84 Da Isoelectric Point: 5.2184
>AVR61311.1 conjugative transfer protein TraI, phosphohydrolase [Proteus mirabilis]
MFKNLFFQAKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTSNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLAMLHIVEADLLFSSNVPSSVRLFSKSEWEATPQTQAEPQSRSSEQPGL
PDASSSIEHGNSSESPSTKSSDQDDELRLASDVNNPQANENAPGDGCENPNNSYDGAISNNVNQHDAEAF
NLPESLAWLPEASSALVMVDEQIMIRYPDAVRHWCAPRKLLAELSRLDWLELDPANPTRKARTVTTKDGV
QEQGLLLKVSISKGLTALIDLPKQDAEPAAAIQNEEALQRPSRTKTTNAQAKEPATRAERKQKPIGPNAN
SSTDPKHAQRQQMVNFVKDLPILLTDGDYPYVDHSADGIRVTIQTLRQVAKEHGIPAGQLLRGISASDEC
QFDEGETVLFTARAER
MFKNLFFQAKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTSNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLAMLHIVEADLLFSSNVPSSVRLFSKSEWEATPQTQAEPQSRSSEQPGL
PDASSSIEHGNSSESPSTKSSDQDDELRLASDVNNPQANENAPGDGCENPNNSYDGAISNNVNQHDAEAF
NLPESLAWLPEASSALVMVDEQIMIRYPDAVRHWCAPRKLLAELSRLDWLELDPANPTRKARTVTTKDGV
QEQGLLLKVSISKGLTALIDLPKQDAEPAAAIQNEEALQRPSRTKTTNAQAKEPATRAERKQKPIGPNAN
SSTDPKHAQRQQMVNFVKDLPILLTDGDYPYVDHSADGIRVTIQTLRQVAKEHGIPAGQLLRGISASDEC
QFDEGETVLFTARAER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16925 | GenBank | AVR61312 |
| Name | traD_-_ICEPmiFra1 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67102.94 Da Isoelectric Point: 6.2511
>AVR61312.1 IncF plasmid conjugative transfer protein TraD [Proteus mirabilis]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYLLYKLQMG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKTNTLAKQAMAMLLFYYERIQPIAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVDNRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYLLYKLQMG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKTNTLAKQAMAMLLFYYERIQPIAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVDNRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16926 | GenBank | AVR61326 |
| Name | traC_-_ICEPmiFra1 |
UniProt ID | _ |
| Length | 799 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 799 a.a. Molecular weight: 90171.75 Da Isoelectric Point: 5.9727
>AVR61326.1 type-IV secretion system protein TraC [Proteus mirabilis]
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTDRELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHIIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTDRELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHIIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 39218..64039
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_27 | 35681..36862 | + | 1182 | AVR61310 | membrane protein | - |
| Locus_28 | 37019..39169 | + | 2151 | AVR61311 | conjugative transfer protein TraI, phosphohydrolase | - |
| Locus_29 | 39218..41038 | + | 1821 | AVR61312 | IncF plasmid conjugative transfer protein TraD | virb4 |
| Locus_30 | 41048..41608 | + | 561 | AVR61313 | conjugative transfer protein | - |
| Locus_31 | 41595..42230 | + | 636 | AVR61314 | conjugal transfer protein | tfc7 |
| Locus_32 | 42302..42940 | + | 639 | AVR61315 | hypothetical protein | - |
| Locus_33 | 42933..44039 | + | 1107 | AVR61316 | zinc peptidase | - |
| Locus_34 | 44176..44457 | + | 282 | AVR61317 | conjugal transfer protein TraL | traL |
| Locus_35 | 44454..45080 | + | 627 | AVR61318 | conjugal transfer pilus assembly protein TraE | traE |
| Locus_36 | 45061..45960 | + | 900 | AVR61319 | conjugal transfer pilus assembly protein TraK | traK |
| Locus_37 | 45963..47252 | + | 1290 | AVR61320 | conjugal transfer pilus assembly protein TraB | traB |
| Locus_38 | 47327..47899 | + | 573 | AVR61321 | conjugal transfer pilus assembly protein TraV | traV |
| Locus_39 | 47896..48282 | + | 387 | AVR61322 | conjugative transfer protein TraA | - |
| Locus_40 | 48461..49294 | + | 834 | AVR61323 | transcriptional regulator with AbiEi antitoxin N-terminal domain | - |
| Locus_41 | 49287..50225 | + | 939 | AVR61324 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| Locus_42 | 50357..51049 | + | 693 | AVR61325 | disulfide isomerase | trbB |
| Locus_43 | 51049..53448 | + | 2400 | AVR61326 | type-IV secretion system protein TraC | virb4 |
| Locus_44 | 53441..53788 | + | 348 | AVR61327 | hypothetical protein | - |
| Locus_45 | 53772..54284 | + | 513 | AVR61328 | conjugative signal peptidase TrhF | - |
| Locus_46 | 54292..55419 | + | 1128 | AVR61329 | IncF plasmid conjugative transfer pilus assembly protein TraW | traW |
| Locus_47 | 55403..56431 | + | 1029 | AVR61330 | conjugal transfer protein TraU | traU |
| Locus_48 | 56434..60126 | + | 3693 | AVR61331 | conjugal transfer protein TraN | traN |
| Locus_49 | 60585..62003 | + | 1419 | AVR61332 | hypothetical protein | - |
| Locus_50 | 62000..64039 | + | 2040 | AVR61333 | ATPase | virb4 |
| Locus_51 | 65164..65730 | - | 567 | AVR61334 | ISL3 family transposase | - |
| Locus_52 | 66473..66985 | - | 513 | AVR61335 | lipoprotein signal peptidase | - |
| Locus_53 | 66989..67885 | - | 897 | AVR61336 | cation transporter | - |
| Locus_54 | 67981..68388 | + | 408 | AVR61337 | MerR family transcriptional regulator | - |
| Locus_55 | 68625..68846 | - | 222 | AVR61338 | hypothetical protein | - |
Host bacterium
| ID | 339 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEPmiFra1 | GenBank | MF490434 |
| Element size | 107395 bp | Coordinate of oriT [Strand] | 4390..4688 [+] |
| Host bacterium | Proteus mirabilis sequence | Coordinate of element | 1..107395 |
Cargo genes
| Drug resistance gene | floR, aph(6)-Id, aph(3'')-Ib, sul2, tet(C), aph(3')-Ia, dfrA32, ere(A), aadA2 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |