Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200380 |
| Name | oriT_ICEPmiChnSTP3 |
| Organism | Proteus mirabilis strain STP3 sequence |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | MT449450 (114477..114775 [-], 299 nt) |
| oriT length | 299 nt |
| IRs (inverted repeats) | 234..242, 254..262 (AAAGCCAAA..TTTGGCTTT) 45..50, 52..57 (CAAACG..CGTTTG) 7..12, 26..31 (TTTGGC..GCCAAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 299 nt
>oriT_ICEPmiChnSTP3
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file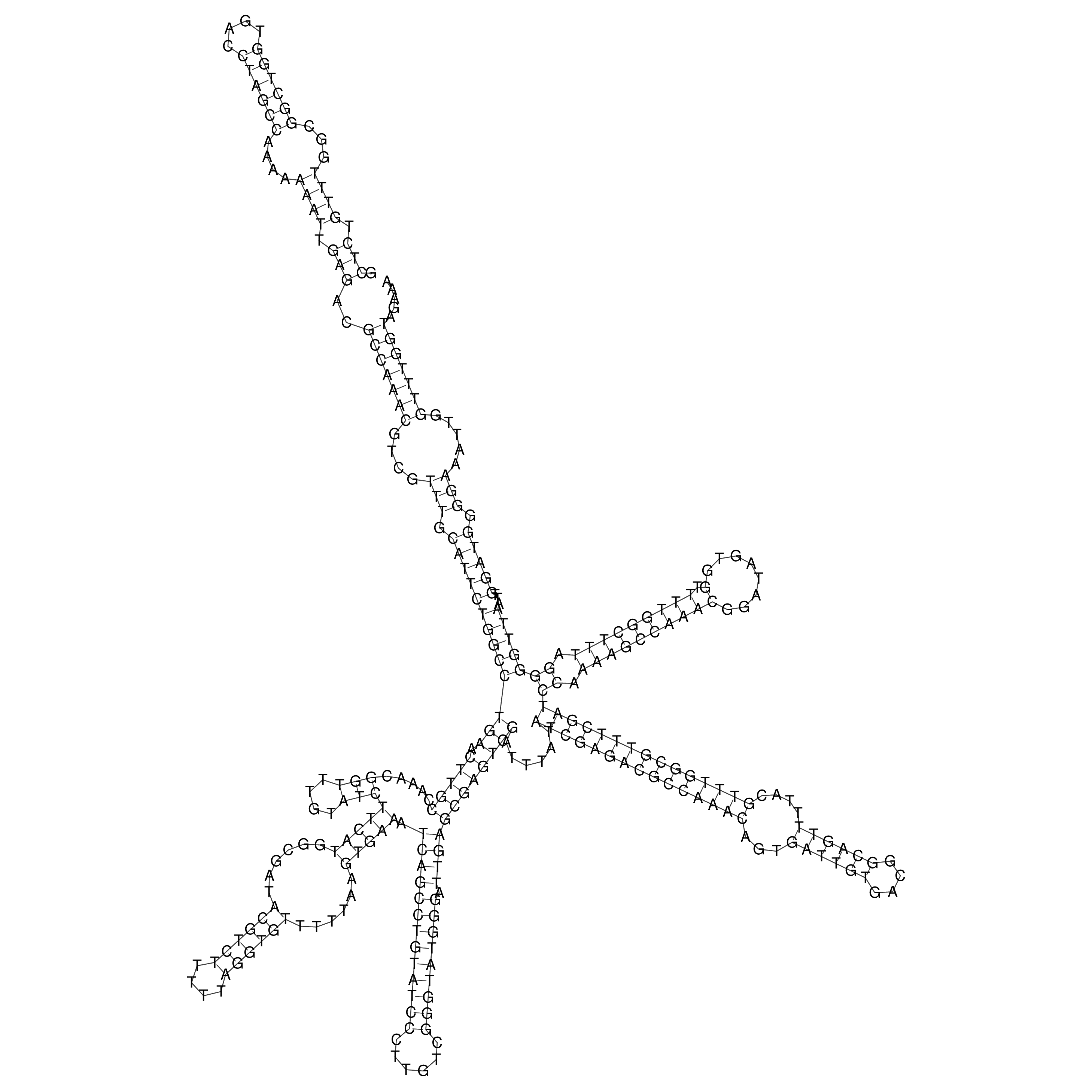
Relaxase
| ID | 14406 | GenBank | QVW29910 |
| Name | traI_-_ICEPmiChnSTP3 |
UniProt ID | _ |
| Length | 716 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 716 a.a. Molecular weight: 79760.84 Da Isoelectric Point: 5.2184
>QVW29910.1 conjugative transfer protein TraI, phosphohydrolase [Proteus mirabilis]
MFKNLFFQAKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTSNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLAMLHIVEADLLFSSNVPSSVRLFSKSEWEATPQTQAEPQSRSSEQPGL
PDASSSIEHGNSSESPSTKSSDQDDELRLASDVNNPQANENAPGDGCENPNNSYDGAISNNVNQHDAEAF
NLPESLAWLPEASSALVMVDEQIMIRYPDAVRHWCAPRKLLAELSRLDWLELDPANPTRKARTVTTKDGV
QEQGLLLKVSISKGLTALIDLPKQDAEPAAAIQNEEALQRPSRTKTTNAQAKEPATRAERKQKPIGPNAN
SSTDPKHAQRQQMVNFVKDLPILLTDGDYPYVDHSADGIRVTIQTLRQVAKEHGIPAGQLLRGISASDEC
QFDEGETVLFTARAER
MFKNLFFQAKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTSNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLAMLHIVEADLLFSSNVPSSVRLFSKSEWEATPQTQAEPQSRSSEQPGL
PDASSSIEHGNSSESPSTKSSDQDDELRLASDVNNPQANENAPGDGCENPNNSYDGAISNNVNQHDAEAF
NLPESLAWLPEASSALVMVDEQIMIRYPDAVRHWCAPRKLLAELSRLDWLELDPANPTRKARTVTTKDGV
QEQGLLLKVSISKGLTALIDLPKQDAEPAAAIQNEEALQRPSRTKTTNAQAKEPATRAERKQKPIGPNAN
SSTDPKHAQRQQMVNFVKDLPILLTDGDYPYVDHSADGIRVTIQTLRQVAKEHGIPAGQLLRGISASDEC
QFDEGETVLFTARAER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16921 | GenBank | QVW29895 |
| Name | traC_-_ICEPmiChnSTP3 |
UniProt ID | _ |
| Length | 799 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 799 a.a. Molecular weight: 90171.75 Da Isoelectric Point: 5.9727
>QVW29895.1 type-IV secretion system protein TraC [Proteus mirabilis]
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTDRELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHIIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTDRELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHIIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16922 | GenBank | QVW29909 |
| Name | traD_-_ICEPmiChnSTP3 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67102.94 Da Isoelectric Point: 6.2511
>QVW29909.1 IncF plasmid conjugative transfer protein TraD [Proteus mirabilis]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYLLYKLQMG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKTNTLAKQAMAMLLFYYERIQPIAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVDNRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYLLYKLQMG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKTNTLAKQAMAMLLFYYERIQPIAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVDNRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 56889..81710
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_61 | 52082..52303 | + | 222 | QVW29883 | hypothetical protein | - |
| Locus_62 | 52540..52947 | - | 408 | QVW29884 | MerR family transcriptional regulator | - |
| Locus_63 | 53043..53939 | + | 897 | QVW29885 | cation transporter | - |
| Locus_64 | 53943..54455 | + | 513 | QVW29886 | lipoprotein signal peptidase | - |
| Locus_65 | 55198..55764 | + | 567 | QVW29887 | ISL3 family transposase | - |
| Locus_66 | 56889..58928 | - | 2040 | QVW29888 | ATPase | virb4 |
| Locus_67 | 58925..60343 | - | 1419 | QVW29889 | hypothetical protein | - |
| Locus_68 | 60802..64494 | - | 3693 | QVW29890 | conjugal transfer protein TraN | traN |
| Locus_69 | 64497..65525 | - | 1029 | QVW29891 | conjugal transfer protein TraU | traU |
| Locus_70 | 65509..66636 | - | 1128 | QVW29892 | IncF plasmid conjugative transfer pilus assembly protein TraW | traW |
| Locus_71 | 66644..67156 | - | 513 | QVW29893 | conjugative signal peptidase TrhF | - |
| Locus_72 | 67140..67487 | - | 348 | QVW29894 | hypothetical protein | - |
| Locus_73 | 67480..69879 | - | 2400 | QVW29895 | type-IV secretion system protein TraC | virb4 |
| Locus_74 | 69879..70571 | - | 693 | QVW29896 | disulfide isomerase | trbB |
| Locus_75 | 70703..71641 | - | 939 | QVW29897 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| Locus_76 | 71634..72467 | - | 834 | QVW29898 | transcriptional regulator with AbiEi antitoxin N-terminal domain | - |
| Locus_77 | 72646..73032 | - | 387 | QVW29899 | conjugative transfer protein TraA | - |
| Locus_78 | 73029..73601 | - | 573 | QVW29900 | conjugal transfer pilus assembly protein TraV | traV |
| Locus_79 | 73676..74965 | - | 1290 | QVW29901 | conjugal transfer pilus assembly protein TraB | traB |
| Locus_80 | 74968..75867 | - | 900 | QVW29902 | conjugal transfer pilus assembly protein TraK | traK |
| Locus_81 | 75848..76240 | - | 393 | QVW29903 | conjugal transfer pilus assembly protein TraE | traE |
| Locus_82 | 76471..76752 | - | 282 | QVW29904 | conjugal transfer protein TraL | traL |
| Locus_83 | 76889..77995 | - | 1107 | QVW29905 | zinc peptidase | - |
| Locus_84 | 77988..78626 | - | 639 | QVW29906 | hypothetical protein | - |
| Locus_85 | 78698..79333 | - | 636 | QVW29907 | conjugal transfer protein | tfc7 |
| Locus_86 | 79320..79880 | - | 561 | QVW29908 | conjugative transfer protein | - |
| Locus_87 | 79890..81710 | - | 1821 | QVW29909 | IncF plasmid conjugative transfer protein TraD | virb4 |
| Locus_88 | 81759..83909 | - | 2151 | QVW29910 | conjugative transfer protein TraI, phosphohydrolase | - |
| Locus_89 | 84066..85247 | - | 1182 | QVW29911 | membrane protein | - |
Host bacterium
| ID | 337 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEPmiChnSTP3 | GenBank | MT449450 |
| Element size | 118916 bp | Coordinate of oriT [Strand] | 114477..114775 [-] |
| Host bacterium | Proteus mirabilis strain STP3 sequence | Coordinate of element | 1..118916 |
Cargo genes
| Drug resistance gene | aadA2, ere(A), dfrA32, cfr, sul1, qacE, ARR-3, catB3, blaOXA-1, aac(6')-Ib-cr, aph(3')-Ia, tet(C), sul2, aph(3'')-Ib, aph(6)-Id, floR |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |