Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200370 |
| Name | oriT_ICEPinKCOM2033-1 |
| Organism | Prevotella intermedia strain KCOM 2033 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP024696 (30068..30264 [-], 197 nt) |
| oriT length | 197 nt |
| IRs (inverted repeats) | 10..17, 20..27 (TATGTTCA..TGAACATA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 197 nt
>oriT_ICEPinKCOM2033-1
ACGGATTTTTATGTTCACCTGAACATAGCAAGATGTCTTTTCAGCCGCTCAAAATATTTTGAGCAGCCACGAAAAGCACTTGCCCTTGCAGGGGGCGGGCTTTCCCTCCGAAGTCGGGAAGCCTTTCAGAACTGCGGTGCTGTAATCCGAAGCGGCGGCTTATGCCGTCAATCCGCTAAATCCAAAGTAATATGAAA
ACGGATTTTTATGTTCACCTGAACATAGCAAGATGTCTTTTCAGCCGCTCAAAATATTTTGAGCAGCCACGAAAAGCACTTGCCCTTGCAGGGGGCGGGCTTTCCCTCCGAAGTCGGGAAGCCTTTCAGAACTGCGGTGCTGTAATCCGAAGCGGCGGCTTATGCCGTCAATCCGCTAAATCCAAAGTAATATGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file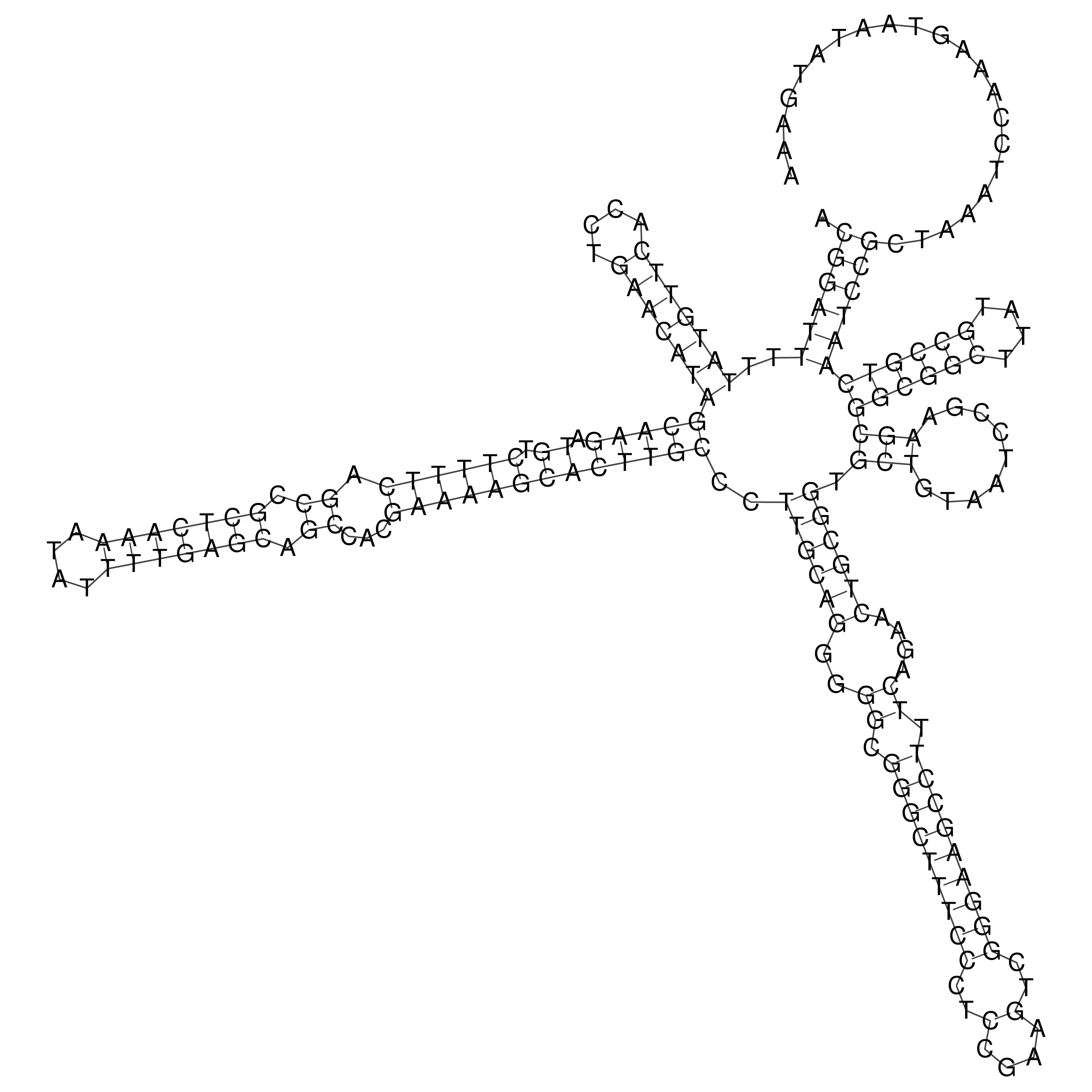
Relaxase
| ID | 14396 | GenBank | ATV53720 |
| Name | Relaxase_CTM50_12240_ICEPinKCOM2033-1 |
UniProt ID | _ |
| Length | 415 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 415 a.a. Molecular weight: 46563.08 Da Isoelectric Point: 9.8931
>ATV53720.1 relaxase [Prevotella intermedia]
MVAKISVGSSLYGAIAYNGEKINEAQGRLLTTNRIYNDGSGTVDIGKAMEGFLTFLPPQMKIEKPVVHIS
LNPHPEDVLTDIELQNIAREYLEKLGFGNQPYLVFKHEDIDRHHLHIVTVNVDENGKRLNRDFLYRRSDR
IRRELEQKYGLHPAERKNQRLDNPLRKVAASAGDVKKQVGNTVKALNGQYRFQTMGEYRALLSLYNMTVE
EARGNVRGREYHGLVYSVTDDKGNKVGNPFKSSLFGKSAGYEAVQKKFVRSKSEIKDRKLADMTKRTVLS
VLQGTYDKDKFVSQLKEKGIDTVLRYTEEGRIYGATFIDHRTGCVLNGSRMGKELSANALQEHFTLPYAG
QPPIPLSIPVDAADKAHGQTAYDSEDISGGMGLLTPEGPAVDAEEEAFIRAMKRKKKKKRKGLGM
MVAKISVGSSLYGAIAYNGEKINEAQGRLLTTNRIYNDGSGTVDIGKAMEGFLTFLPPQMKIEKPVVHIS
LNPHPEDVLTDIELQNIAREYLEKLGFGNQPYLVFKHEDIDRHHLHIVTVNVDENGKRLNRDFLYRRSDR
IRRELEQKYGLHPAERKNQRLDNPLRKVAASAGDVKKQVGNTVKALNGQYRFQTMGEYRALLSLYNMTVE
EARGNVRGREYHGLVYSVTDDKGNKVGNPFKSSLFGKSAGYEAVQKKFVRSKSEIKDRKLADMTKRTVLS
VLQGTYDKDKFVSQLKEKGIDTVLRYTEEGRIYGATFIDHRTGCVLNGSRMGKELSANALQEHFTLPYAG
QPPIPLSIPVDAADKAHGQTAYDSEDISGGMGLLTPEGPAVDAEEEAFIRAMKRKKKKKRKGLGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16904 | GenBank | ATV53719 |
| Name | t4cp2_CTM50_12235_ICEPinKCOM2033-1 |
UniProt ID | _ |
| Length | 672 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 672 a.a. Molecular weight: 77086.10 Da Isoelectric Point: 7.1001
>ATV53719.1 conjugal transfer protein TraG [Prevotella intermedia]
MSQQEDDLRALAKIMDFLRAVSIILVVMNVYWFCYEAIRLWGVNIGVVDKILLNFDRTAGLFHSILYTKL
FSVLLLALSCLGTKGVKGEKITWGRIWTAFAVGFVLFFLNWWLLPLPLPLEAVTGLYVLTIGTGYVCLLM
GGLWMSRLLKHNLMEDVFNNENESFMQETRLIESEYSVNLPTRFYYRKRWNNGWINVVNPFRASIVLGTP
GSGKSYAVVNNFIKQQIEKGFSQYIYDFKYPDLSTIAYNHLLNHPDGYKVKPKFYVINFDDPRRSHRCNP
IHPDFMEDITDAYESAYTIMLNLNKTWVQKQGDFFVESPIILFASIIWYLKIYQNGKFCTFPHAIEFLNR
RYEDIFPILTSYPELENYLSPFMDAWLGGAAEQLMGQIASAKIPLSRMISPQLYWVMSDSEFTLDINNPE
EPKILCVGNNPDRQNIYGAALGLYNSRIVKLINKKGMLKSSVIIDELPTIYFKGLDNLIATARSNKVAVC
LGFQDFSQLVRDYGDKEAKVVMNTVGNIFSGQVVGETAKTLSERFGKVLQKRQSISINRQDVSTSINTQM
DALIPPSKISGLTQGMFVGSVSDNFNERIEQKIFHCEIVVDAEKVKREESAYKKIPVITNFTDEDGNDRM
KETVQANYRRIKEEVKQIVQEELERIKNDPVLCKLLPDNETV
MSQQEDDLRALAKIMDFLRAVSIILVVMNVYWFCYEAIRLWGVNIGVVDKILLNFDRTAGLFHSILYTKL
FSVLLLALSCLGTKGVKGEKITWGRIWTAFAVGFVLFFLNWWLLPLPLPLEAVTGLYVLTIGTGYVCLLM
GGLWMSRLLKHNLMEDVFNNENESFMQETRLIESEYSVNLPTRFYYRKRWNNGWINVVNPFRASIVLGTP
GSGKSYAVVNNFIKQQIEKGFSQYIYDFKYPDLSTIAYNHLLNHPDGYKVKPKFYVINFDDPRRSHRCNP
IHPDFMEDITDAYESAYTIMLNLNKTWVQKQGDFFVESPIILFASIIWYLKIYQNGKFCTFPHAIEFLNR
RYEDIFPILTSYPELENYLSPFMDAWLGGAAEQLMGQIASAKIPLSRMISPQLYWVMSDSEFTLDINNPE
EPKILCVGNNPDRQNIYGAALGLYNSRIVKLINKKGMLKSSVIIDELPTIYFKGLDNLIATARSNKVAVC
LGFQDFSQLVRDYGDKEAKVVMNTVGNIFSGQVVGETAKTLSERFGKVLQKRQSISINRQDVSTSINTQM
DALIPPSKISGLTQGMFVGSVSDNFNERIEQKIFHCEIVVDAEKVKREESAYKKIPVITNFTDEDGNDRM
KETVQANYRRIKEEVKQIVQEELERIKNDPVLCKLLPDNETV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2736069..2746624
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CTM50_12240 | 2731198..2732445 | - | 1248 | ATV53720 | relaxase | - |
| CTM50_12245 | 2732424..2732852 | - | 429 | ATV53721 | MobA protein | - |
| CTM50_12250 | 2732849..2733073 | - | 225 | ATV53945 | hypothetical protein | - |
| CTM50_12255 | 2733122..2733289 | - | 168 | ATV53722 | hypothetical protein | - |
| CTM50_12260 | 2733274..2734323 | + | 1050 | ATV53723 | ParA family protein | - |
| CTM50_12265 | 2734326..2734766 | + | 441 | ATV53946 | hypothetical protein | - |
| CTM50_12270 | 2734769..2735122 | + | 354 | ATV53724 | conjugal transfer protein TraB | - |
| CTM50_12275 | 2735139..2735867 | + | 729 | ATV53725 | conjugal transfer protein TraD | - |
| CTM50_12280 | 2736069..2736386 | + | 318 | ATV53726 | conjugal transfer protein TraE | traE |
| CTM50_12285 | 2736397..2736729 | + | 333 | ATV53727 | conjugal transfer protein TraF | traF |
| CTM50_12290 | 2736726..2739230 | + | 2505 | ATV53728 | conjugal transfer protein TraG | virb4 |
| CTM50_12295 | 2739271..2739651 | + | 381 | ATV53729 | conjugal transfer protein TraH | traH |
| CTM50_12300 | 2739675..2740304 | + | 630 | ATV53730 | hypothetical protein | traI |
| CTM50_12305 | 2740308..2741312 | + | 1005 | ATV53731 | conjugative transposon protein TraJ | traJ |
| CTM50_12310 | 2741344..2741967 | + | 624 | ATV53732 | conjugative transposon protein TraK | traK |
| CTM50_12315 | 2741974..2742282 | + | 309 | ATV53733 | conjugal transfer protein TraL | traL |
| CTM50_12320 | 2742263..2743615 | + | 1353 | ATV53734 | conjugative transposon protein TraM | traM |
| CTM50_12325 | 2743650..2744636 | + | 987 | ATV53735 | conjugative transposon protein TraN | traN |
| CTM50_12330 | 2744639..2745214 | + | 576 | ATV53736 | conjugal transfer protein TraO | traO |
| CTM50_12335 | 2745222..2746124 | + | 903 | ATV53737 | DNA primase | traP |
| CTM50_12340 | 2746121..2746624 | + | 504 | ATV53738 | conjugal transfer protein TraQ | traQ |
| CTM50_12345 | 2746621..2747148 | + | 528 | ATV53739 | lysozyme | - |
| CTM50_12350 | 2747357..2747602 | + | 246 | ATV53947 | hypothetical protein | - |
| CTM50_12355 | 2747636..2747944 | - | 309 | ATV53948 | hypothetical protein | - |
| CTM50_12360 | 2747998..2748243 | - | 246 | ATV53740 | hypothetical protein | - |
| CTM50_12365 | 2748240..2748473 | - | 234 | ATV53741 | DUF3873 domain-containing protein | - |
| CTM50_12370 | 2748493..2748750 | - | 258 | ATV53742 | hypothetical protein | - |
| CTM50_12375 | 2748775..2750106 | - | 1332 | ATV53743 | hypothetical protein | - |
| CTM50_12380 | 2750103..2750519 | - | 417 | ATV53744 | PcfK-like protein | - |
| CTM50_12385 | 2750533..2750754 | - | 222 | ATV53950 | hypothetical protein | - |
| CTM50_12390 | 2750766..2750981 | - | 216 | ATV53949 | hypothetical protein | - |
| CTM50_12395 | 2750887..2751131 | + | 245 | Protein_2416 | hypothetical protein | - |
| CTM50_12400 | 2751143..2751553 | - | 411 | ATV53745 | hypothetical protein | - |
Host bacterium
| ID | 327 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEPinKCOM2033-1 | GenBank | CP024696 |
| Element size | 2777805 bp | Coordinate of oriT [Strand] | 30068..30264 [-] |
| Host bacterium | Prevotella intermedia strain KCOM 2033 | Coordinate of element | 2702780..2752400 |
Cargo genes
| Drug resistance gene | tet(Q) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9 |