Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200365 |
| Name | oriT_ICEPciChn180 |
| Organism | Proteus terrae subsp. cibarius strain SDQ8C180-2T |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP073356 (120336..120634 [-], 299 nt) |
| oriT length | 299 nt |
| IRs (inverted repeats) | 234..242, 254..262 (AAAGCCAAA..TTTGGCTTT) 45..50, 52..57 (CAAACG..CGTTTG) 7..12, 26..31 (TTTGGC..GCCAAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 299 nt
>oriT_ICEPciChn180
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file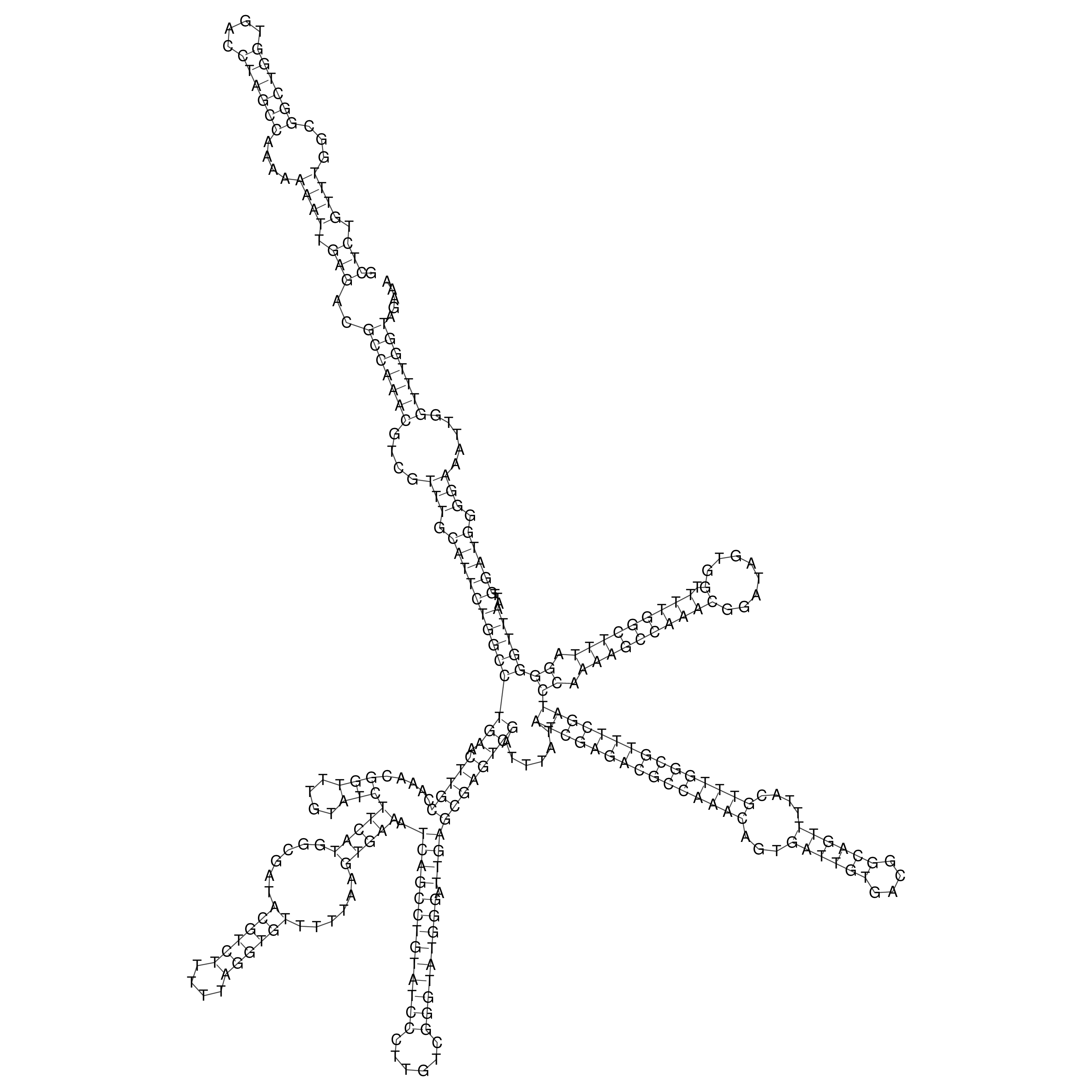
Relaxase
| ID | 14391 | GenBank | UAX01280 |
| Name | TraI_2_KDN45_14915_ICEPciChn180 |
UniProt ID | _ |
| Length | 716 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 716 a.a. Molecular weight: 79644.48 Da Isoelectric Point: 5.3431
>UAX01280.1 TraI domain-containing protein [Proteus terrae subsp. cibarius]
MFKNLFFQAKAIPELSSQLDADIPRYPPFLKGLPAASPKDLQSTQDELIAKLRQVLGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLPMLHISEADLLFSSNVPSSVRLFSKSEWEATQQTQAESQSRSSEHSDL
PEASSSIEHGNSSESPSTNSSEQDDELSHASDVNHLQATENAPGDGCEKPNNSYDGAISNNVNQHDAEAL
NLTESLVWLPEASSALVMVGEQILIRYPDAVRPWCAPRKLLAELSRLDWLELDPANPTRKARTVTTNDGV
HEQGLLLKVSISKGLTALIDFSKQDAESVTAIQNEEASQRPSRTETANAQAKELATRAERKQKPIAPNAN
SSTDPKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANAHGIPAGQLLRGISASDEC
QFDEGETVLFTAHAKR
MFKNLFFQAKAIPELSSQLDADIPRYPPFLKGLPAASPKDLQSTQDELIAKLRQVLGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLPMLHISEADLLFSSNVPSSVRLFSKSEWEATQQTQAESQSRSSEHSDL
PEASSSIEHGNSSESPSTNSSEQDDELSHASDVNHLQATENAPGDGCEKPNNSYDGAISNNVNQHDAEAL
NLTESLVWLPEASSALVMVGEQILIRYPDAVRPWCAPRKLLAELSRLDWLELDPANPTRKARTVTTNDGV
HEQGLLLKVSISKGLTALIDFSKQDAESVTAIQNEEASQRPSRTETANAQAKELATRAERKQKPIAPNAN
SSTDPKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANAHGIPAGQLLRGISASDEC
QFDEGETVLFTAHAKR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16895 | GenBank | UAX01267 |
| Name | traC_KDN45_14845_ICEPciChn180 |
UniProt ID | _ |
| Length | 799 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 799 a.a. Molecular weight: 90171.75 Da Isoelectric Point: 5.9735
>UAX01267.1 type IV secretion system protein TraC [Proteus terrae subsp. cibarius]
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16896 | GenBank | UAX01279 |
| Name | traD_KDN45_14910_ICEPciChn180 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67059.86 Da Isoelectric Point: 6.5728
>UAX01279.1 conjugative transfer system coupling protein TraD [Proteus terrae subsp. cibarius]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLHKLQKG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPSFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLHKLQKG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPSFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3238187..3258052
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KDN45_14810 | 3233762..3235666 | - | 1905 | UAX01260 | ATP-dependent helicase | - |
| KDN45_14815 | 3235675..3237627 | - | 1953 | UAX01261 | AAA family ATPase | - |
| KDN45_14820 | 3238187..3241879 | - | 3693 | UAX01262 | conjugal transfer protein TraN | traN |
| KDN45_14825 | 3241882..3242910 | - | 1029 | UAX01263 | TraU family protein | traU |
| KDN45_14830 | 3242894..3244018 | - | 1125 | UAX01264 | pilus assembly protein | traW |
| KDN45_14835 | 3244029..3244541 | - | 513 | UAX01265 | S26 family signal peptidase | - |
| KDN45_14840 | 3244525..3244872 | - | 348 | UAX01266 | plasmid-related protein | - |
| KDN45_14845 | 3244865..3247264 | - | 2400 | UAX01267 | type IV secretion system protein TraC | virb4 |
| KDN45_14850 | 3247264..3247956 | - | 693 | UAX01268 | DsbC family protein | trbB |
| KDN45_14855 | 3248088..3249026 | - | 939 | UAX01269 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| KDN45_14860 | 3249019..3249810 | - | 792 | UAX01270 | type IV toxin-antitoxin system AbiEi family antitoxin | - |
| KDN45_14865 | 3250031..3250417 | - | 387 | UAX01271 | hypothetical protein | - |
| KDN45_14870 | 3250414..3250986 | - | 573 | UAX01272 | type IV conjugative transfer system lipoprotein TraV | traV |
| KDN45_14875 | 3251061..3252350 | - | 1290 | UAX01273 | TraB/VirB10 family protein | traB |
| KDN45_14880 | 3252353..3253249 | - | 897 | UAX03602 | type-F conjugative transfer system secretin TraK | traK |
| KDN45_14885 | 3253233..3253859 | - | 627 | UAX01274 | pilus assembly protein | traE |
| KDN45_14890 | 3253856..3254137 | - | 282 | UAX01275 | type IV conjugative transfer system protein TraL | traL |
| KDN45_14895 | 3254426..3255013 | + | 588 | UAX01276 | plasmid-related protein | - |
| KDN45_14900 | 3255040..3255675 | - | 636 | UAX01277 | DUF4400 domain-containing protein | tfc7 |
| KDN45_14905 | 3255662..3256222 | - | 561 | UAX01278 | conjugative transfer protein | - |
| KDN45_14910 | 3256232..3258052 | - | 1821 | UAX01279 | conjugative transfer system coupling protein TraD | virb4 |
| KDN45_14915 | 3258100..3260250 | - | 2151 | UAX01280 | TraI domain-containing protein | - |
| KDN45_14920 | 3260368..3261081 | - | 714 | UAX01281 | hypothetical protein | - |
| KDN45_14925 | 3261082..3262272 | - | 1191 | UAX01282 | nucleotidyltransferase | - |
Host bacterium
| ID | 322 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEPciChn180 | GenBank | CP073356 |
| Element size | 3959345 bp | Coordinate of oriT [Strand] | 120336..120634 [-] |
| Host bacterium | Proteus terrae subsp. cibarius strain SDQ8C180-2T | Coordinate of element | 3209019..3335275 |
Cargo genes
| Drug resistance gene | sul2, tet(X6), aph(4)-Ia, aac(3)-IVa, aph(3')-Ia, aph(3'')-Ib, aph(6)-Id |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIA |