Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200359 |
| Name | oriT_ICEMfrJAM7 |
| Organism | Methylophaga frappieri strain JAM7 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP003380 (107477..107775 [-], 299 nt) |
| oriT length | 299 nt |
| IRs (inverted repeats) | 45..50, 52..57 (CAAACG..CGTTTG) 7..12, 26..31 (TTTGGC..GCCAAA) 1..8, 20..27 (GCTCTGTT..AACAGAGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 299 nt
>oriT_ICEMfrJAM7
GCTCTGTTTGGCGGCGGATAACAGAGCCAAAAAAATCGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTCGCCAAACGGTTTGTATCTTCATGACGATACGTCTTTTTAGGCGTTTTTAAGTGAAATCGGGCTGTATCCCTTGTCAGGTATGGGATTGCGCGAGTTGATTTATATCGAGACGCCAAACAGTGATTGTTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
GCTCTGTTTGGCGGCGGATAACAGAGCCAAAAAAATCGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTCGCCAAACGGTTTGTATCTTCATGACGATACGTCTTTTTAGGCGTTTTTAAGTGAAATCGGGCTGTATCCCTTGTCAGGTATGGGATTGCGCGAGTTGATTTATATCGAGACGCCAAACAGTGATTGTTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file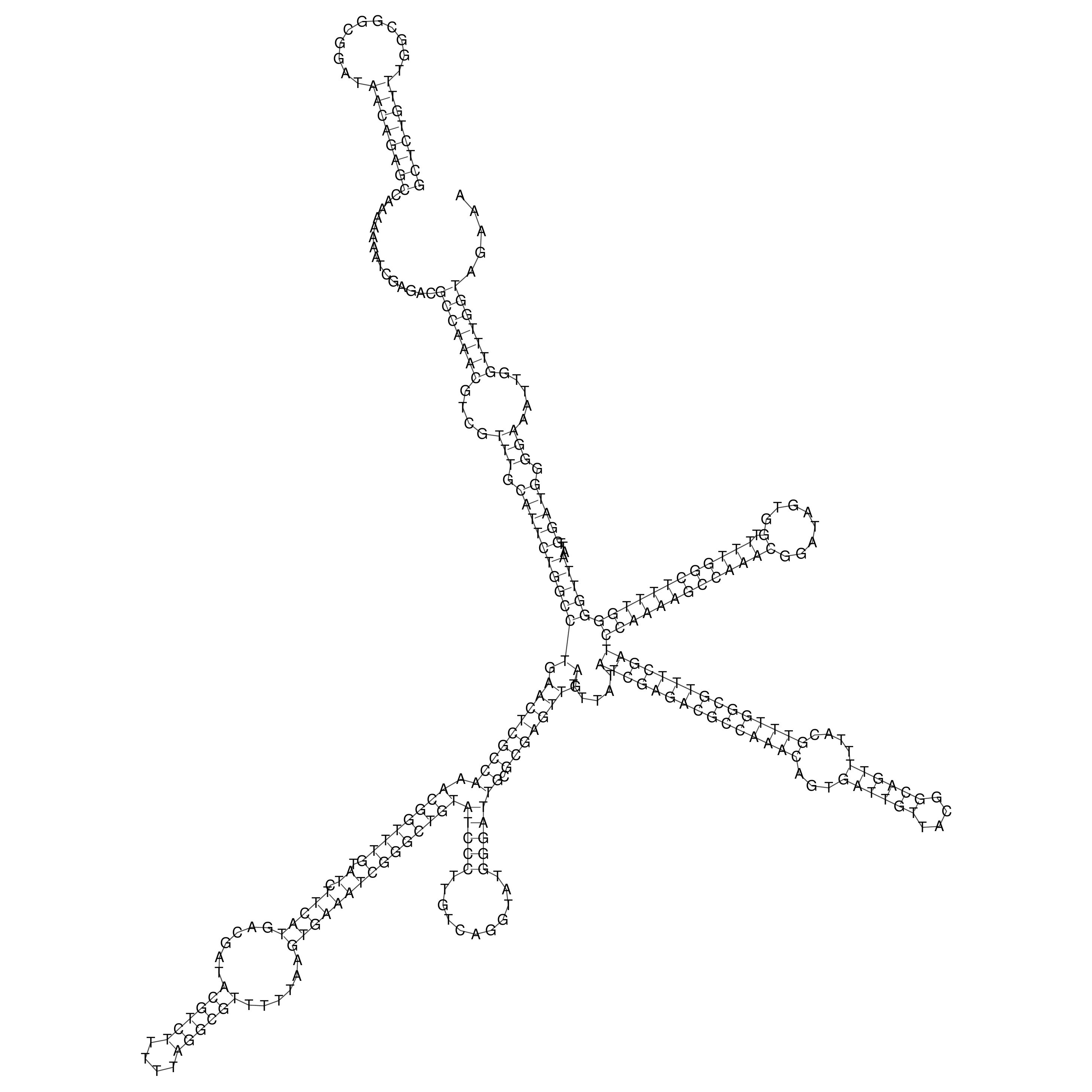
Relaxase
| ID | 14387 | GenBank | AFJ01717 |
| Name | TraI_2_Q7C_544_ICEMfrJAM7 |
UniProt ID | _ |
| Length | 716 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 716 a.a. Molecular weight: 79624.66 Da Isoelectric Point: 5.2822
>AFJ01717.1 Conjugative transfer protein TraI, relaxase [Methylophaga frappieri]
MFKNLFFQTKALPELSSQPDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNQREFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVTSGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPDHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWVRKSNQSTHLYLVWKSAAKDIIELLSKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLPMLHISEADLLFSSNVPSSVRLFSKSEWEATQQTQAEPQSRSSEHPDL
PEASSSIEHSNSTKSPSTKPSEQDDELRLASDVNNPQATENAPGDGCEKPNDSNDAAIGNNVSQQNEATL
NLPESLAWLPGASSALVMVGEQILIRYPDAVRPWCAPRKLLAELSQLDWLELDPANPTRKARTVTTKDGV
QEQGLLLKVSISKGLTALIDLPKQDAESVSAIQNKEASQRPSRTETTSAQAKELATRAERKQKPIAANAN
SSTDPKHVQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAER
MFKNLFFQTKALPELSSQPDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNQREFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVTSGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPDHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWVRKSNQSTHLYLVWKSAAKDIIELLSKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLPMLHISEADLLFSSNVPSSVRLFSKSEWEATQQTQAEPQSRSSEHPDL
PEASSSIEHSNSTKSPSTKPSEQDDELRLASDVNNPQATENAPGDGCEKPNDSNDAAIGNNVSQQNEATL
NLPESLAWLPGASSALVMVGEQILIRYPDAVRPWCAPRKLLAELSQLDWLELDPANPTRKARTVTTKDGV
QEQGLLLKVSISKGLTALIDLPKQDAESVSAIQNKEASQRPSRTETTSAQAKELATRAERKQKPIAANAN
SSTDPKHVQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16887 | GenBank | AFJ01702 |
| Name | t4cp2_Q7C_529_ICEMfrJAM7 |
UniProt ID | _ |
| Length | 799 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 799 a.a. Molecular weight: 90187.75 Da Isoelectric Point: 5.9735
>AFJ01702.1 IncF plasmid conjugative transfer pilus assembly protein TraC [Methylophaga frappieri]
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRSIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRSIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16888 | GenBank | AFJ01716 |
| Name | t4cp2_Q7C_543_ICEMfrJAM7 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67126.95 Da Isoelectric Point: 6.5130
>AFJ01716.1 IncF plasmid conjugative transfer protein TraD [Methylophaga frappieri]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQKG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPIAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQKG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPIAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 562388..582154
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Q7C_522 | 557410..560463 | + | 3054 | AFJ01695 | putative glycosyltransferase | - |
| Q7C_523 | 561185..562066 | - | 882 | AFJ01696 | molybdenum cofactor biosynthesis enzyme | - |
| Q7C_524 | 562388..566080 | - | 3693 | AFJ01697 | IncF plasmid conjugative transfer protein TraN | traN |
| Q7C_525 | 566083..567111 | - | 1029 | AFJ01698 | IncF plasmid conjugative transfer pilus assembly protein TraU | traU |
| Q7C_526 | 567095..568219 | - | 1125 | AFJ01699 | IncF plasmid conjugative transfer pilus assembly protein TraW | traW |
| Q7C_527 | 568230..568742 | - | 513 | AFJ01700 | Conjugative signal peptidase TrhF | - |
| Q7C_528 | 568726..569073 | - | 348 | AFJ01701 | Conjugative transfer protein 345 | - |
| Q7C_529 | 569066..571465 | - | 2400 | AFJ01702 | IncF plasmid conjugative transfer pilus assembly protein TraC | virb4 |
| Q7C_530 | 571465..572157 | - | 693 | AFJ01703 | Thiol:disulfide involved in conjugative transfer | trbB |
| Q7C_531 | 572289..573227 | - | 939 | AFJ01704 | Ync | - |
| Q7C_532 | 573220..574053 | - | 834 | AFJ01705 | Ynd | - |
| Q7C_533 | 574231..574617 | - | 387 | AFJ01706 | Conjugative transfer protein TraA | - |
| Q7C_534 | 574614..575264 | - | 651 | AFJ01707 | Conjugative transfer protein TraV | traV |
| Q7C_535 | 575261..576550 | - | 1290 | AFJ01708 | IncF plasmid conjugative transfer pilus assembly protein TraB | traB |
| Q7C_536 | 576550..577449 | - | 900 | AFJ01709 | IncF plasmid conjugative transfer pilus assembly protein TraK | traK |
| Q7C_537 | 577433..578059 | - | 627 | AFJ01710 | IncF plasmid conjugative transfer pilus assembly protein TraE | traE |
| Q7C_538 | 578056..578337 | - | 282 | AFJ01711 | IncF plasmid conjugative transfer pilus assembly protein TraL | traL |
| Q7C_539 | 578518..578841 | + | 324 | AFJ01712 | phage-like protein | - |
| Q7C_540 | 578838..579116 | + | 279 | AFJ01713 | putative transcriptional regulator with C-terminal CBS domains | - |
| Q7C_541 | 579142..579777 | - | 636 | AFJ01714 | Conjugative transfer protein s043 | tfc7 |
| Q7C_542 | 579764..580324 | - | 561 | AFJ01715 | Conjugative transfer protein 234 | - |
| Q7C_543 | 580334..582154 | - | 1821 | AFJ01716 | IncF plasmid conjugative transfer protein TraD | virb4 |
| Q7C_544 | 582203..584353 | - | 2151 | AFJ01717 | Conjugative transfer protein TraI, relaxase | - |
| Q7C_545 | 584434..585831 | - | 1398 | AFJ01718 | hypothetical protein | - |
Host bacterium
| ID | 316 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEMfrJAM7 | GenBank | CP003380 |
| Element size | 2697465 bp | Coordinate of oriT [Strand] | 107477..107775 [-] |
| Host bacterium | Methylophaga frappieri strain JAM7 | Coordinate of element | 495596..606053 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |