Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200356 |
| Name | oriT_ICEGEN |
| Organism | Vibrio cholerae strain IDH_4268 sequence |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | MK165650 (4841..5139 [+], 299 nt) |
| oriT length | 299 nt |
| IRs (inverted repeats) | 234..242, 254..262 (AAAGCCAAA..TTTGGCTTT) 45..50, 52..57 (CAAACG..CGTTTG) 7..12, 26..31 (TTTGGC..GCCAAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 299 nt
>oriT_ICEGEN
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file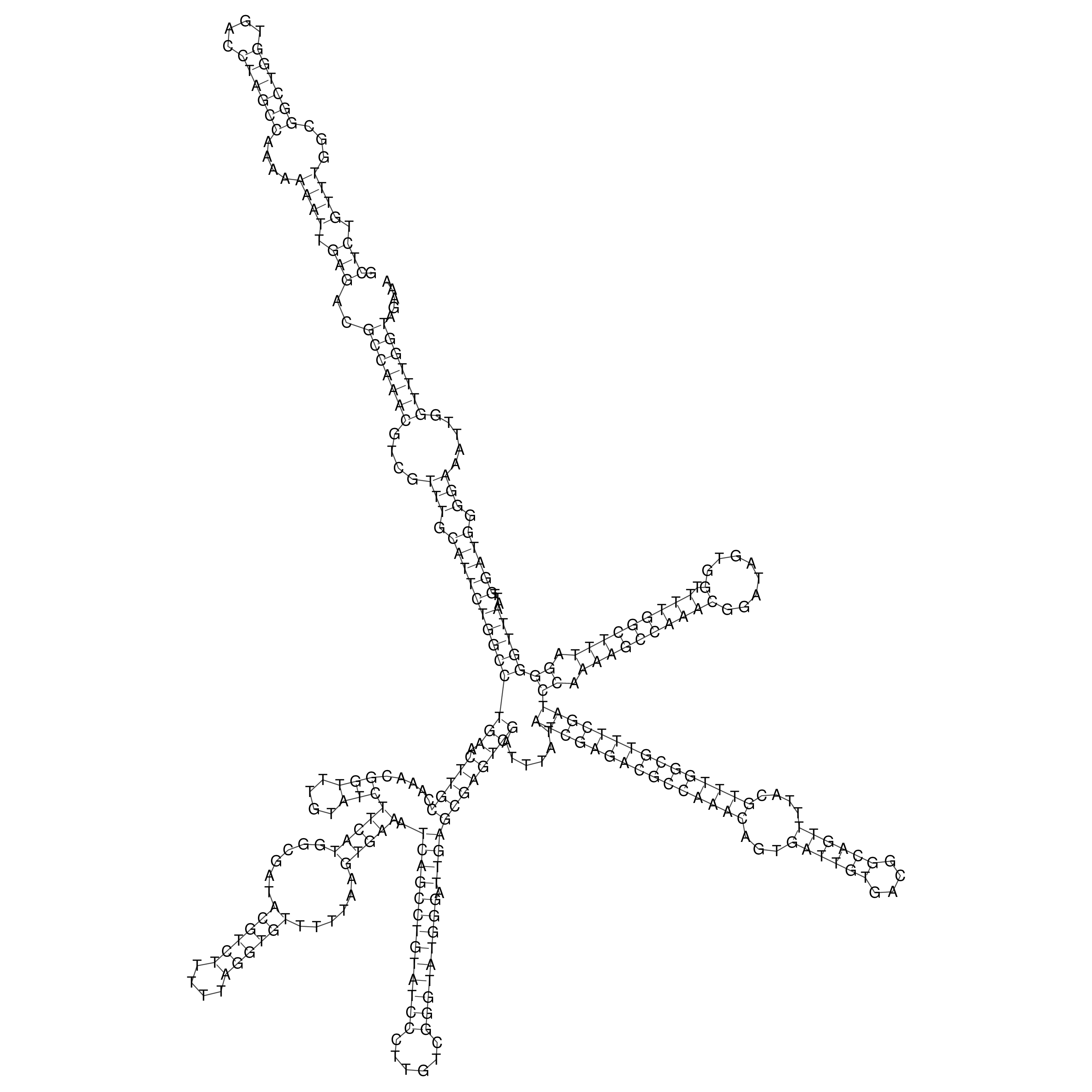
Relaxase
| ID | 14385 | GenBank | QAY18328 |
| Name | traI_-_ICEGEN |
UniProt ID | _ |
| Length | 716 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 716 a.a. Molecular weight: 80010.08 Da Isoelectric Point: 5.6098
>QAY18328.1 conjugal transfer pilus assembly protein TraI [Vibrio cholerae]
MFKNLFFQAKALPDLSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNLRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWATNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWLRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLPMLHISEADLLFSSNVPSSVRLFSKSEWEATQQTQAEPQSRSSEHSDL
PEASSSIEHRKSTESPSTNSSEQDDELRHASDVNHLEENENVPGDGYEKPNNSYDGAISNNVNQHDAEAL
NLPESLVWLPEASSALVMVGEQVLIRYPDAVRHWCAPRKLLAELSRLDWLELDPANPTRKARTVTTKGGV
QEQGLLLKVSISKGLTSLIDLPKQDAESAAAIQNEEASQRPSRTKTTNAQAREPAKRAERKQKPIAPNAN
SSTDPKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVAKEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAER
MFKNLFFQAKALPDLSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNLRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWATNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWLRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDKPIWLPMLHISEADLLFSSNVPSSVRLFSKSEWEATQQTQAEPQSRSSEHSDL
PEASSSIEHRKSTESPSTNSSEQDDELRHASDVNHLEENENVPGDGYEKPNNSYDGAISNNVNQHDAEAL
NLPESLVWLPEASSALVMVGEQVLIRYPDAVRHWCAPRKLLAELSRLDWLELDPANPTRKARTVTTKGGV
QEQGLLLKVSISKGLTSLIDLPKQDAESAAAIQNEEASQRPSRTKTTNAQAREPAKRAERKQKPIAPNAN
SSTDPKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVAKEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16879 | GenBank | QAY18329 |
| Name | traD_-_ICEGEN |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67115.98 Da Isoelectric Point: 6.5130
>QAY18329.1 conjugal transfer pilus assembly protein TraD [Vibrio cholerae]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQMG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKKTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQMG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKKTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16880 | GenBank | QAY18343 |
| Name | traC_-_ICEGEN |
UniProt ID | _ |
| Length | 799 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 799 a.a. Molecular weight: 90170.73 Da Isoelectric Point: 5.9735
>QAY18343.1 conjugal transfer ATP-binding protein TraC [Vibrio cholerae]
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVNEMTDRNWLAALSAQLNWGKDASWRNPSPVRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVNEMTDRNWLAALSAQLNWGKDASWRNPSPVRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 43242..63107
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_32 | 39301..40071 | + | 771 | QAY18326 | hypothetical protein | - |
| Locus_33 | 40079..40921 | + | 843 | QAY18327 | abortive phage resistance protein | - |
| Locus_34 | 41043..43193 | + | 2151 | QAY18328 | conjugal transfer pilus assembly protein TraI | - |
| Locus_35 | 43242..45062 | + | 1821 | QAY18329 | conjugal transfer pilus assembly protein TraD | virb4 |
| Locus_36 | 45072..45632 | + | 561 | QAY18330 | conjugative transfer protein 234 | - |
| Locus_37 | 45610..46254 | + | 645 | QAY18331 | conjugative transfer protein | tfc7 |
| Locus_38 | 46281..46868 | - | 588 | QAY18332 | DNA helicase family protein | - |
| Locus_39 | 46882..47121 | - | 240 | QAY18333 | plasmid related protein | - |
| Locus_40 | 47157..47438 | + | 282 | QAY18334 | conjugative transfer pilus assembly protein TraL | traL |
| Locus_41 | 47435..48061 | + | 627 | QAY18335 | conjugative transfer pilus assembly protein TraE | traE |
| Locus_42 | 48042..48941 | + | 900 | QAY18336 | conjugal transfer pilus assembly protein TraK | traK |
| Locus_43 | 48944..50233 | + | 1290 | QAY18337 | conjugal transfer pilus assembly protein TraB | traB |
| Locus_44 | 50230..50880 | + | 651 | QAY18338 | Conjugative transfer protein TraV | traV |
| Locus_45 | 50877..51263 | + | 387 | QAY18339 | conjugal transfer pilus assembly protein TraA | - |
| Locus_46 | 51442..52275 | + | 834 | QAY18340 | Ynd | - |
| Locus_47 | 52268..53206 | + | 939 | QAY18341 | Ync | - |
| Locus_48 | 53338..54030 | + | 693 | QAY18342 | disulfide isomerase DsbC | trbB |
| Locus_49 | 54030..56429 | + | 2400 | QAY18343 | conjugal transfer ATP-binding protein TraC | virb4 |
| Locus_50 | 56422..56769 | + | 348 | QAY18344 | hypothetical protein | - |
| Locus_51 | 56753..57265 | + | 513 | QAY18345 | conjugative signal peptidase TrhF | - |
| Locus_52 | 57273..58400 | + | 1128 | QAY18346 | conjugal transfer pilus assembly protein TraW | traW |
| Locus_53 | 58384..59412 | + | 1029 | QAY18347 | conjugal transfer pilus assembly protein TraU | traU |
| Locus_54 | 59415..63107 | + | 3693 | QAY18348 | conjugal transfer mating pair stabilization protein TraN | traN |
| Locus_55 | 63198..64814 | - | 1617 | QAY18349 | SMC domain protein | - |
| Locus_56 | 64894..65649 | - | 756 | QAY18350 | ATP binding domain IstB | - |
| Locus_57 | 65639..67147 | - | 1509 | QAY18351 | integrase catalytic subunit IstA | - |
Host bacterium
| ID | 313 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEGEN | GenBank | MK165650 |
| Element size | 96718 bp | Coordinate of oriT [Strand] | 4841..5139 [+] |
| Host bacterium | Vibrio cholerae strain IDH_4268 sequence | Coordinate of element | 1..96718 |
Cargo genes
| Drug resistance gene | floR, aph(6)-Id, aph(3'')-Ib, sul2, dfrA1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |