Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200355 |
| Name | oriT_ICEErZJ |
| Organism | Erysipelothrix rhusiopathiae sequence |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | MG812141 (61942..62075 [-], 134 nt) |
| oriT length | 134 nt |
| IRs (inverted repeats) | 66..71, 84..89 (AAATCC..GGATTT) 6..12, 24..30 (ACCCCCC..GGGGGGT) 7..12, 23..28 (CCCCCC..GGGGGG) |
| Location of nic site | 76..77 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 134 nt
ACTTAACCCCCCGTATCTAACAGGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file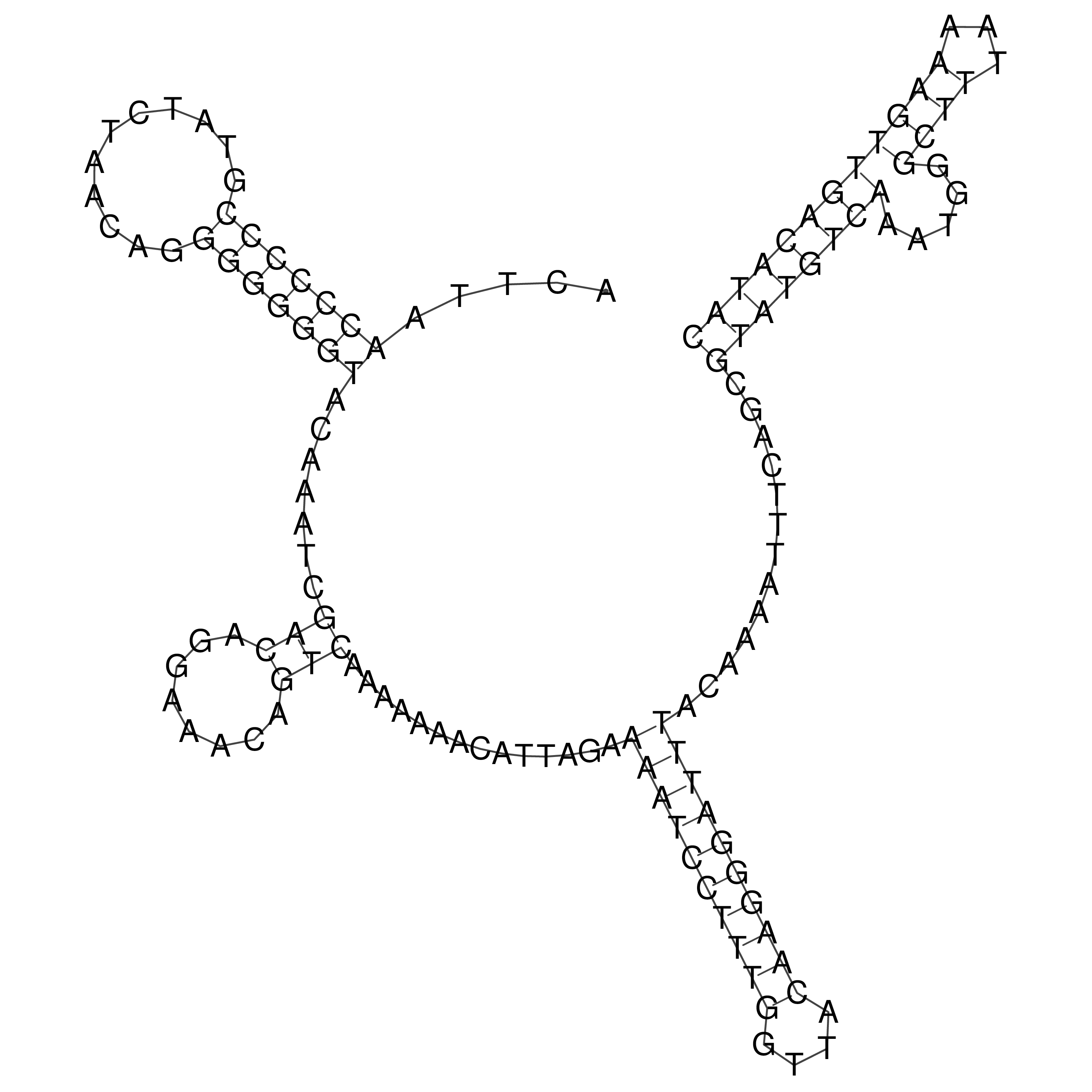
Relaxase
| ID | 14383 | GenBank | AYC07713 |
| Name | Rep_trans_Er0106_065_ICEErZJ |
UniProt ID | _ |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14384 | GenBank | AYC07720 |
| Name | Relaxase_Er0106_072_ICEErZJ |
UniProt ID | _ |
| Length | 444 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 444 a.a. Molecular weight: 52649.16 Da Isoelectric Point: 9.7582
MAITKIHPIKSTLNLAIDYITKSEKTDEKILISSFKCHPSTAHIQFMKTREDNDTTGKVLARHLIQSFLP
GEVDPIKAHEIGMELCKKILKEDYEFVLATHIDRGHIHNHIIFNNVNYKTGKCYQSNKKTYHKIRYQSDE
LCKENKLSVIDKYYEAYKRKYKTAGKSWYEYDQNKKGNSWKSKLQFDIDRMINKSKSWEEFLENMKSIDY
EIKFGKHIAFRHKDKQRFTRAKTIGEDYAEEKIKERIDLAIKNKANPIKKRVGNVIDISTNKKAQSSKGY
EVWARKHNIKTMADSIIKLREQGINSITQLDDLIKKSADDRQDLLDKIKKIETEMKSLSQDMENINTINK
YREIYKYHKKNPEDKQFAEEYYSDLSVYKIAAKEILENYNKLPNTKEILSNLDKLQEKKNTLMQEYSLNK
EQFSDLVQYRKNYENYYGKEVERT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7884 | GenBank | AYC07701 |
| Name | AYC07701_ICEErZJ |
UniProt ID | _ |
| Length | 405 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 405 a.a. Molecular weight: 47021.92 Da Isoelectric Point: 9.7978
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16878 | GenBank | AYC07714 |
| Name | tcpA_Er0106_066_ICEErZJ |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53414.28 Da Isoelectric Point: 8.9454
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVDLGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 8338..15353
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Er0106_006 | 3811..4095 | + | 285 | AYC07656 | Phage antirepressor protein | - |
| Er0106_007 | 4174..4368 | + | 195 | AYC07734 | Phage antirepressor protein | - |
| Er0106_008 | 4531..4773 | + | 243 | AYC07657 | TrsK-like protein | - |
| Er0106_009 | 4879..5127 | + | 249 | AYC07658 | TrsK-like protein | - |
| Er0106_010 | 5210..5422 | + | 213 | AYC07659 | TrsK-like protein | - |
| Er0106_011 | 5422..5625 | + | 204 | AYC07660 | TrsK-like protein | - |
| Er0106_012 | 5626..5847 | + | 222 | AYC07661 | TrsK-like protein;relaxase/mobilization protein | - |
| Er0106_013 | 6228..6527 | + | 300 | AYC07662 | hypothetical protein | - |
| Er0106_014 | 6550..6843 | + | 294 | AYC07663 | hypothetical protein | - |
| Er0106_015 | 7367..7534 | + | 168 | AYC07664 | AraC family transcriptional regulator | - |
| Er0106_016 | 7632..7805 | + | 174 | AYC07665 | AraC family transcriptional regulator | - |
| Er0106_017 | 7850..8068 | + | 219 | AYC07666 | AraC family transcriptional regulator | - |
| Er0106_018 | 8338..8649 | + | 312 | AYC07667 | single-stranded DNA-binding protein | cd424 |
| Er0106_019 | 8651..8866 | + | 216 | AYC07668 | conjugal transfer protein | prgF |
| Er0106_020 | 8877..9740 | + | 864 | AYC07669 | membrane protein | prgHa |
| Er0106_021 | 9750..10040 | + | 291 | AYC07670 | hypothetical protein | - |
| Er0106_022 | 10042..10452 | + | 411 | AYC07671 | conjugal transfer protein | prgIa |
| Er0106_023 | 10523..12766 | + | 2244 | AYC07672 | type IV secretory pathway protein VirB4 | virb4 |
| Er0106_024 | 12774..15353 | + | 2580 | AYC07673 | NLP/P60 family protein | cd419b |
| Er0106_025 | 15633..17162 | + | 1530 | AYC07674 | Intracellular protein transport protein USO1 | - |
| Er0106_026 | 17177..18043 | + | 867 | AYC07675 | mature parasite-infected erythrocyte surface antigen | - |
| Er0106_027 | 18107..19828 | + | 1722 | AYC07676 | DNA topoisomerase III | - |
Region 2: 52829..64240
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Er0106_053 | 48016..48219 | - | 204 | AYC07702 | Xis-Tn protein | - |
| Er0106_054 | 48680..48910 | - | 231 | AYC07703 | hypothetical protein | - |
| Er0106_055 | 48907..49329 | - | 423 | AYC07704 | hypothetical protein | - |
| Er0106_056 | 49834..50187 | + | 354 | AYC07705 | DNA-binding protein | - |
| Er0106_057 | 50533..52452 | - | 1920 | AYC07706 | Tetracycline resistance protein TetM | - |
| Er0106_058 | 52829..53761 | - | 933 | AYC07707 | conjugal transfer protein | orf13 |
| Er0106_059 | 53758..54759 | - | 1002 | AYC07708 | lipoprotein, NLP/P60 family | orf14 |
| Er0106_060 | 54756..56867 | - | 2112 | AYC07709 | membrane protein | orf15 |
| Er0106_061 | 56936..59383 | - | 2448 | AYC07710 | ATP/GTP-binding protein | virb4 |
| Er0106_062 | 59367..59759 | - | 393 | AYC07711 | membrane protein | orf17a |
| Er0106_063 | 59848..60345 | - | 498 | AYC07712 | Antirestriction protein | - |
| Er0106_064 | 60462..60683 | - | 222 | AYC07735 | Conjugation related protein | orf19 |
| Er0106_065 | 60726..61931 | - | 1206 | AYC07713 | Tn916, transcriptional regulator | - |
| Er0106_066 | 62110..63495 | - | 1386 | AYC07714 | Tn916, transcriptional regulator | virb4 |
| Er0106_067 | 63524..63910 | - | 387 | AYC07715 | conjugal transfer protein | orf23 |
| Er0106_068 | 63926..64240 | - | 315 | AYC07716 | conjugal transfer protein | orf23 |
| Er0106_069 | 64604..65239 | + | 636 | AYC07717 | hypothetical protein | - |
| Er0106_070 | 65251..65772 | + | 522 | AYC07718 | hypothetical protein | - |
| Er0106_071 | 65769..66626 | + | 858 | AYC07719 | hypothetical protein | - |
| Er0106_072 | 66646..67980 | - | 1335 | AYC07720 | Rlx-like protein | - |
| Er0106_073 | 67982..68338 | - | 357 | AYC07721 | hypothetical protein | - |
Host bacterium
| ID | 312 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEErZJ | GenBank | MG812141 |
| Element size | 79087 bp | Coordinate of oriT [Strand] | 61942..62075 [-] |
| Host bacterium | Erysipelothrix rhusiopathiae sequence | Coordinate of element | 1..79087 |
Cargo genes
| Drug resistance gene | ant(6)-Ia, lsa(E), lnu(B), aph(3')-III, tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |