Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200349 |
| Name | oriT_ICEBcaI48-1 |
| Organism | Bacteroides caecimuris strain I48 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP015401 (33204..33400 [-], 197 nt) |
| oriT length | 197 nt |
| IRs (inverted repeats) | 10..17, 20..27 (TATGTTCA..TGAACATA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 197 nt
>oriT_ICEBcaI48-1
ACGGATTTTTATGTTCACCTGAACATAGCAAGATGTCTTTTCAGCCGCTCAAAATATTTTGAGCAGCCACGAAAAGCACTTGCCCTTGCAGGGGGCGGGCTTTCCCTCCGAAGTCGGGAAGCCTTTCAGAACTGCGGTGCTGTAATCCGAAGCGGCGGCTTATGCCGTCAATCCGCTAAATCCAAAGTAATATGAAA
ACGGATTTTTATGTTCACCTGAACATAGCAAGATGTCTTTTCAGCCGCTCAAAATATTTTGAGCAGCCACGAAAAGCACTTGCCCTTGCAGGGGGCGGGCTTTCCCTCCGAAGTCGGGAAGCCTTTCAGAACTGCGGTGCTGTAATCCGAAGCGGCGGCTTATGCCGTCAATCCGCTAAATCCAAAGTAATATGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file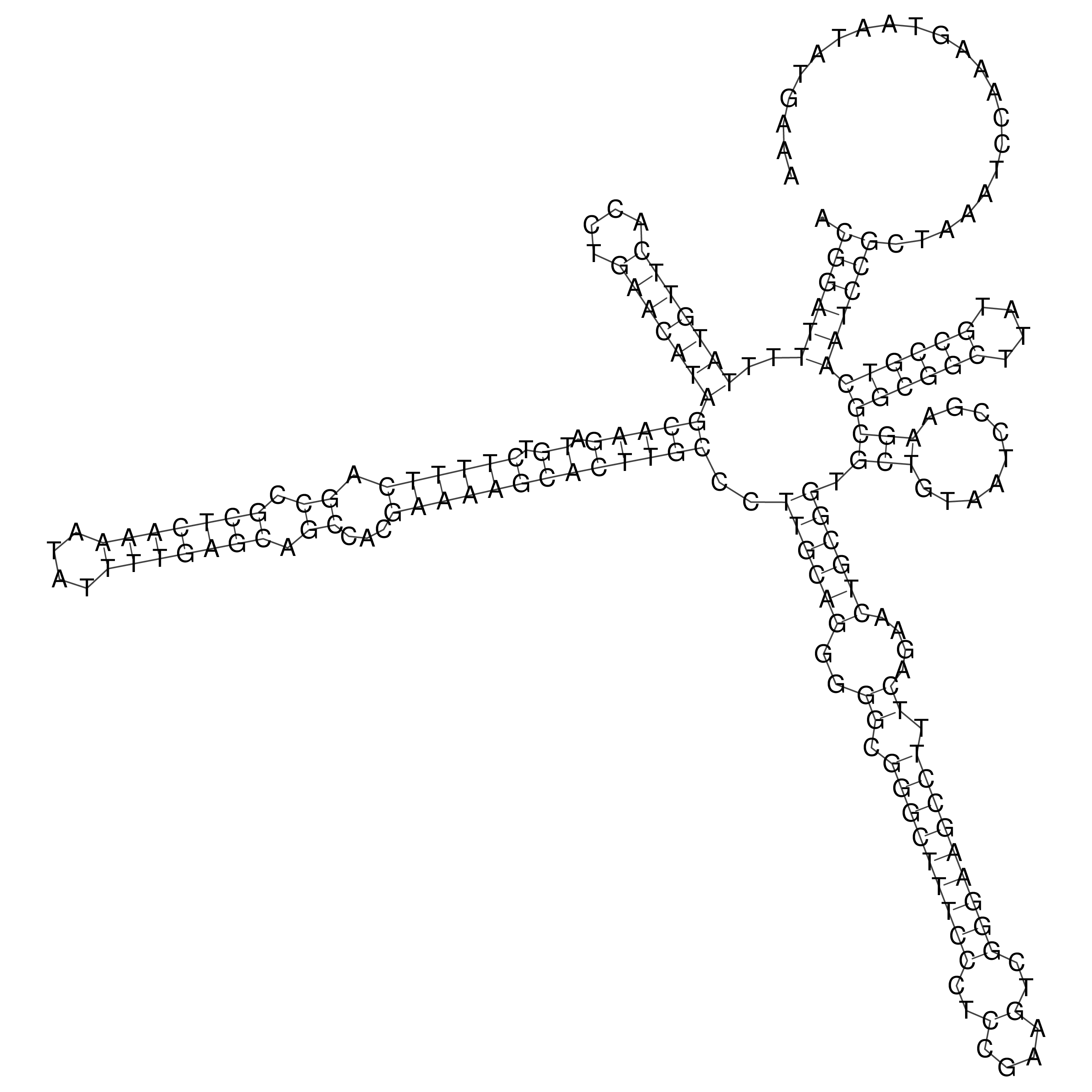
Relaxase
| ID | 14374 | GenBank | ANU57185 |
| Name | Relaxase_A4V03_06090_ICEBcaI48-1 |
UniProt ID | _ |
| Length | 415 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 415 a.a. Molecular weight: 46563.08 Da Isoelectric Point: 9.8931
>ANU57185.1 relaxase [Bacteroides caecimuris]
MVAKISVGSSLYGAIAYNGEKINEAQGRLLTTNRIYNDGSGTVDIGKAMEGFLTFLPPQMKIEKPVVHIS
LNPHPEDVLTDIELQNIAREYLEKLGFGNQPYLVFKHEDIDRHHLHIVTVNVDENGKRLNRDFLYRRSDR
IRRELEQKYGLHPAERKNQRLDNPLRKVAASAGDVKKQVGNTVKALNGQYRFQTMGEYRALLSLYNMTVE
EARGNVRGREYHGLVYSVTDDKGNKVGNPFKSSLFGKSAGYEAVQKKFVRSKSEIKDRKLADMTKRTVLS
VLQGTYDKDKFVSQLKEKGIDTVLRYTEEGRIYGATFIDHRTGCVLNGSRMGKELSANALQEHFTLPYAG
QPPIPLSIPVDAADKAHGQTAYDSEDISGGMGLLTPEGPAVDAEEEAFIRAMKRKKKKKRKGLGM
MVAKISVGSSLYGAIAYNGEKINEAQGRLLTTNRIYNDGSGTVDIGKAMEGFLTFLPPQMKIEKPVVHIS
LNPHPEDVLTDIELQNIAREYLEKLGFGNQPYLVFKHEDIDRHHLHIVTVNVDENGKRLNRDFLYRRSDR
IRRELEQKYGLHPAERKNQRLDNPLRKVAASAGDVKKQVGNTVKALNGQYRFQTMGEYRALLSLYNMTVE
EARGNVRGREYHGLVYSVTDDKGNKVGNPFKSSLFGKSAGYEAVQKKFVRSKSEIKDRKLADMTKRTVLS
VLQGTYDKDKFVSQLKEKGIDTVLRYTEEGRIYGATFIDHRTGCVLNGSRMGKELSANALQEHFTLPYAG
QPPIPLSIPVDAADKAHGQTAYDSEDISGGMGLLTPEGPAVDAEEEAFIRAMKRKKKKKRKGLGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16869 | GenBank | ANU57184 |
| Name | t4cp2_A4V03_06085_ICEBcaI48-1 |
UniProt ID | _ |
| Length | 672 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 672 a.a. Molecular weight: 77086.10 Da Isoelectric Point: 7.1001
>ANU57184.1 conjugal transfer protein TraG [Bacteroides caecimuris]
MSQQEDDLRALAKIMDFLRAVSIILVVMNVYWFCYEAIRLWGVNIGVVDKILLNFDRTAGLFHSILYTKL
FSVLLLALSCLGTKGVKGEKITWGRIWTAFAVGFVLFFLNWWLLPLPLPLEAVTGLYVLTIGTGYVCLLM
GGLWMSRLLKHNLMEDVFNNENESFMQETRLIESEYSVNLPTRFYYRKRWNNGWINVVNPFRASIVLGTP
GSGKSYAVVNNFIKQQIEKGFSQYIYDFKYPDLSTIAYNHLLNHPDGYKVKPKFYVINFDDPRRSHRCNP
IHPDFMEDITDAYESAYTIMLNLNKTWVQKQGDFFVESPIILFASIIWYLKIYQNGKFCTFPHAIEFLNR
RYEDIFPILTSYPELENYLSPFMDAWLGGAAEQLMGQIASAKIPLSRMISPQLYWVMSDSEFTLDINNPE
EPKILCVGNNPDRQNIYGAALGLYNSRIVKLINKKGMLKSSVIIDELPTIYFKGLDNLIATARSNKVAVC
LGFQDFSQLVRDYGDKEAKVVMNTVGNIFSGQVVGETAKTLSERFGKVLQKRQSISINRQDVSTSINTQM
DALIPPSKISGLTQGMFVGSVSDNFNERIEQKIFHCEIVVDAEKVKREESAYKKIPVITNFTDEDGNDRM
KETVQANYRRIKEEVKQIVQEELERIKNDPVLCKLLPDNETV
MSQQEDDLRALAKIMDFLRAVSIILVVMNVYWFCYEAIRLWGVNIGVVDKILLNFDRTAGLFHSILYTKL
FSVLLLALSCLGTKGVKGEKITWGRIWTAFAVGFVLFFLNWWLLPLPLPLEAVTGLYVLTIGTGYVCLLM
GGLWMSRLLKHNLMEDVFNNENESFMQETRLIESEYSVNLPTRFYYRKRWNNGWINVVNPFRASIVLGTP
GSGKSYAVVNNFIKQQIEKGFSQYIYDFKYPDLSTIAYNHLLNHPDGYKVKPKFYVINFDDPRRSHRCNP
IHPDFMEDITDAYESAYTIMLNLNKTWVQKQGDFFVESPIILFASIIWYLKIYQNGKFCTFPHAIEFLNR
RYEDIFPILTSYPELENYLSPFMDAWLGGAAEQLMGQIASAKIPLSRMISPQLYWVMSDSEFTLDINNPE
EPKILCVGNNPDRQNIYGAALGLYNSRIVKLINKKGMLKSSVIIDELPTIYFKGLDNLIATARSNKVAVC
LGFQDFSQLVRDYGDKEAKVVMNTVGNIFSGQVVGETAKTLSERFGKVLQKRQSISINRQDVSTSINTQM
DALIPPSKISGLTQGMFVGSVSDNFNERIEQKIFHCEIVVDAEKVKREESAYKKIPVITNFTDEDGNDRM
KETVQANYRRIKEEVKQIVQEELERIKNDPVLCKLLPDNETV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1485237..1498281
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| A4V03_06090 | 1480366..1481613 | - | 1248 | ANU57185 | relaxase | - |
| A4V03_06095 | 1481592..1482020 | - | 429 | ANU57186 | MobA protein | - |
| A4V03_20340 | 1482017..1482241 | - | 225 | ARE60540 | hypothetical protein | - |
| A4V03_06100 | 1482442..1483491 | + | 1050 | ANU59708 | conjugal transfer protein TraA | - |
| A4V03_06105 | 1483494..1483934 | + | 441 | ANU59706 | hypothetical protein | - |
| A4V03_06110 | 1483937..1484290 | + | 354 | ANU57187 | conjugal transfer protein TraB | - |
| A4V03_06115 | 1484307..1485035 | + | 729 | ANU59707 | conjugal transfer protein TraD | - |
| A4V03_06120 | 1485237..1485554 | + | 318 | ANU57188 | conjugal transfer protein TraE | traE |
| A4V03_06125 | 1485565..1485897 | + | 333 | ANU57189 | conjugal transfer protein TraF | traF |
| A4V03_06130 | 1485894..1488398 | + | 2505 | ANU57190 | conjugal transfer protein TraG | virb4 |
| A4V03_06135 | 1488439..1488819 | + | 381 | ANU57191 | conjugal transfer protein TraH | traH |
| A4V03_06140 | 1488861..1489472 | + | 612 | ANU57192 | hypothetical protein | traI |
| A4V03_06145 | 1489476..1490480 | + | 1005 | ANU57193 | conjugative transposon protein TraJ | traJ |
| A4V03_06150 | 1490512..1491135 | + | 624 | ANU57194 | conjugative transposon protein TraK | traK |
| A4V03_06155 | 1491142..1491450 | + | 309 | ANU57195 | conjugal transfer protein TraL | traL |
| A4V03_06160 | 1491431..1492783 | + | 1353 | ANU57196 | conjugative transposon protein TraM | traM |
| A4V03_06165 | 1492818..1493243 | + | 426 | ARE60481 | conjugal transfer protein TraN | traN |
| A4V03_20345 | 1493262..1493750 | + | 489 | ARE60482 | hypothetical protein | - |
| A4V03_06170 | 1493785..1495596 | + | 1812 | ANU57197 | group II intron reverse transcriptase/maturase | - |
| A4V03_06175 | 1495568..1496293 | + | 726 | ANU57198 | conjugative transposon protein TraN | traN |
| A4V03_06180 | 1496296..1496871 | + | 576 | ANU57199 | conjugal transfer protein TraO | traO |
| A4V03_06185 | 1496879..1497781 | + | 903 | ANU57200 | DNA primase | traP |
| A4V03_06190 | 1497778..1498281 | + | 504 | ANU57201 | conjugal transfer protein TraQ | traQ |
| A4V03_06195 | 1498278..1498805 | + | 528 | ANU57202 | lysozyme | - |
| A4V03_06200 | 1498870..1499259 | + | 390 | ANU57203 | hypothetical protein | - |
| A4V03_06205 | 1499293..1499601 | - | 309 | ANU59709 | hypothetical protein | - |
| A4V03_06210 | 1499655..1499900 | - | 246 | ANU57204 | hypothetical protein | - |
| A4V03_06215 | 1499897..1500130 | - | 234 | ANU57205 | hypothetical protein | - |
| A4V03_06220 | 1500150..1500407 | - | 258 | ANU57206 | hypothetical protein | - |
| A4V03_06225 | 1500432..1501763 | - | 1332 | ANU57207 | hypothetical protein | - |
| A4V03_06230 | 1501760..1502176 | - | 417 | ANU57208 | PcfK-like protein | - |
| A4V03_06235 | 1502190..1502411 | - | 222 | ANU57209 | hypothetical protein | - |
| A4V03_06240 | 1502423..1502638 | - | 216 | ANU57210 | hypothetical protein | - |
| A4V03_20350 | 1502544..1502788 | + | 245 | Protein_1258 | hypothetical protein | - |
| A4V03_06245 | 1502935..1503198 | + | 264 | ANU57211 | hypothetical protein | - |
Host bacterium
| ID | 306 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEBcaI48-1 | GenBank | CP015401 |
| Element size | 4839927 bp | Coordinate of oriT [Strand] | 33204..33400 [-] |
| Host bacterium | Bacteroides caecimuris strain I48 | Coordinate of element | 1448812..1503529 |
Cargo genes
| Drug resistance gene | tet(Q) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9 |