Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200341 |
| Name | oriT_ICE_SsuZYH33_Tn916 |
| Organism | Streptococcus suis 05ZYH33 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_009442 (30698..30830 [-], 133 nt) |
| oriT length | 133 nt |
| IRs (inverted repeats) | 65..70, 83..88 (AAATCC..GGATTT) 6..12, 23..29 (ACCCCCC..GGGGGGT) |
| Location of nic site | 75..76 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 133 nt
>oriT_ICE_SsuZYH33_Tn916
ACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
ACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file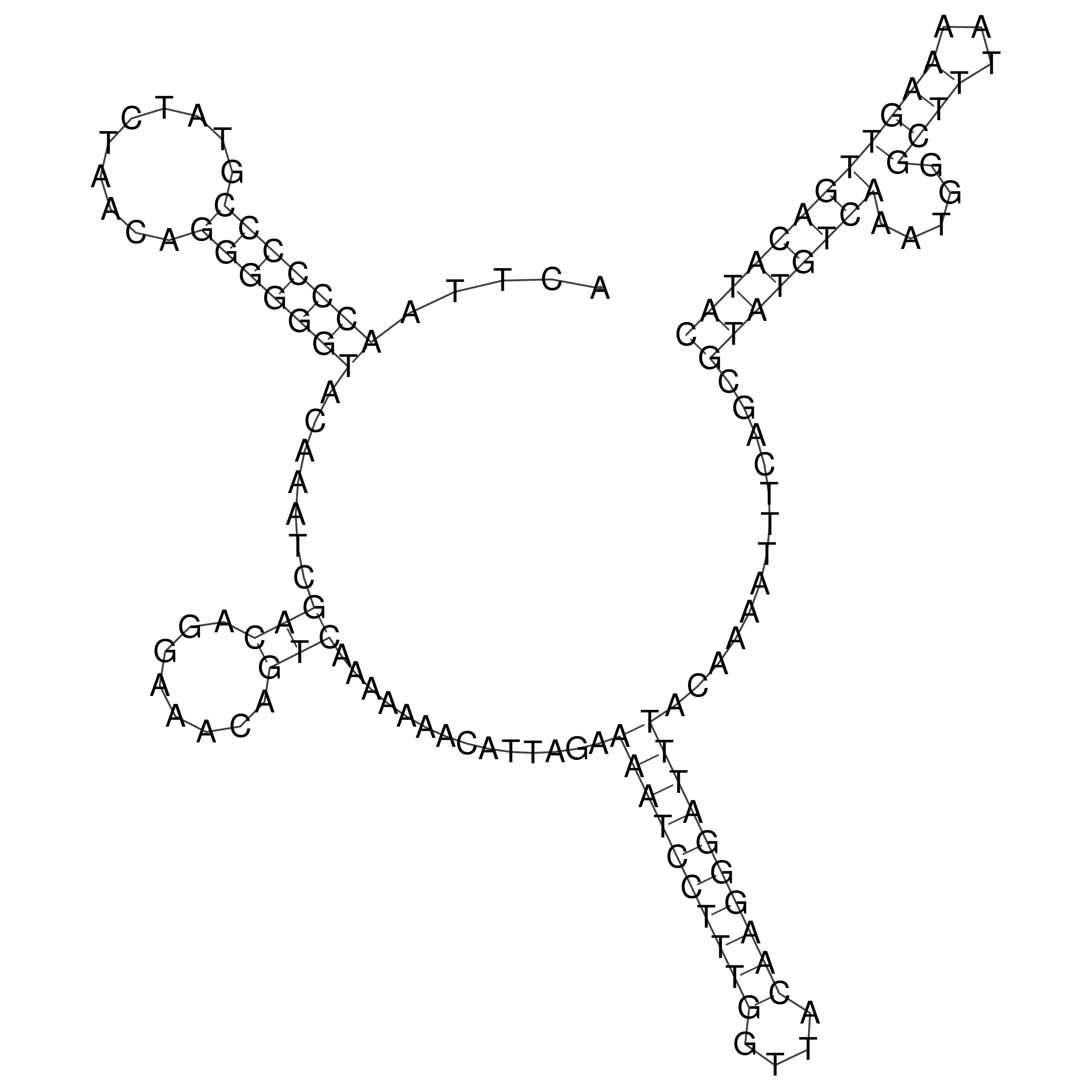
Relaxase
| ID | 14365 | GenBank | YP_001198296 |
| Name | Rep_trans_SSU05_0930_ICE_SsuZYH33_Tn916 |
UniProt ID | _ |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
>YP_001198296.1 Tn916, transcriptional regulator [Streptococcus suis 05ZYH33]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7882 | GenBank | YP_001198283 |
| Name | YP_001198283_ICE_SsuZYH33_Tn916 |
UniProt ID | _ |
| Length | 405 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 405 a.a. Molecular weight: 47021.92 Da Isoelectric Point: 9.7978
>YP_001198283.1 Tn916, transposase [Streptococcus suis 05ZYH33]
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDVVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16856 | GenBank | YP_001198297 |
| Name | tcpA_SSU05_0931_ICE_SsuZYH33_Tn916 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>YP_001198297.1 hypothetical protein SSU05_0931 [Streptococcus suis 05ZYH33]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 894536..914673
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SSU05_0918 | 889724..889927 | - | 204 | YP_001198284 | hypothetical protein | - |
| SSU05_0919 | 890615..891046 | - | 432 | YP_001198285 | hypothetical protein | - |
| SSU05_0920 | 891541..891894 | + | 354 | YP_001198286 | Tn916, transcriptional regulator | - |
| SSU05_0922 | 892240..894174 | - | 1935 | YP_001198287 | translation elongation factor (GTPases) | - |
| SSU05_0921 | 893067..893219 | + | 153 | YP_001198288 | hypothetical protein | - |
| SSU05_0923 | 894536..895468 | - | 933 | YP_001198289 | Tn916 hypothetical protein | orf13 |
| SSU05_0924 | 895465..896466 | - | 1002 | YP_001198290 | hypothetical protein | orf14 |
| SSU05_0925 | 896463..898640 | - | 2178 | YP_001198291 | hypothetical protein | orf15 |
| SSU05_0926 | 898643..901099 | - | 2457 | YP_001198292 | hypothetical protein | virb4 |
| SSU05_0927 | 901074..901580 | - | 507 | YP_001198293 | hypothetical protein | orf17a |
| SSU05_0928 | 901555..902052 | - | 498 | YP_001198294 | hypothetical protein | - |
| SSU05_0929 | 902169..902390 | - | 222 | YP_001198295 | hypothetical protein | orf19 |
| SSU05_0930 | 902433..903638 | - | 1206 | YP_001198296 | Tn916, transcriptional regulator | - |
| SSU05_0931 | 903816..905201 | - | 1386 | YP_001198297 | hypothetical protein | virb4 |
| SSU05_0932 | 905230..905616 | - | 387 | YP_001198298 | hypothetical protein | orf23 |
| SSU05_0933 | 905632..905946 | - | 315 | YP_001198299 | hypothetical protein | orf23 |
| SSU05_0934 | 907006..908550 | - | 1545 | YP_001198300 | DNA helicase | - |
| SSU05_0935 | 908507..910594 | - | 2088 | YP_001198301 | hypothetical protein | - |
| SSU05_0936 | 910591..911367 | - | 777 | YP_001198302 | Signal recognition particle GTPase | - |
| SSU05_0937 | 911367..911873 | - | 507 | YP_001198303 | transcriptional regulator | - |
| SSU05_0938 | 911913..912266 | - | 354 | YP_001198304 | ATPases with chaperone activity, ATP-binding subunit | - |
| SSU05_0939 | 912253..912642 | - | 390 | YP_001198305 | methyl-accepting chemotaxis protein | gbs1346 |
| SSU05_0940 | 912639..912866 | - | 228 | YP_001198306 | hypothetical protein | gbs1347 |
| SSU05_0941 | 912920..913996 | - | 1077 | YP_001198307 | DNA primase (type) | - |
| SSU05_0942 | 914035..914673 | - | 639 | YP_001198308 | hypothetical protein | prgL |
| SSU05_0943 | 916074..916673 | - | 600 | YP_001198309 | DNA-binding response regulator | - |
| SSU05_0944 | 916651..917841 | - | 1191 | YP_001198310 | two-component sensor histidine kinase | - |
| SSU05_0945 | 918568..918852 | - | 285 | YP_001198311 | hypothetical protein | - |
Region 2: 894536..959143
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SSU05_0918 | 889724..889927 | - | 204 | YP_001198284 | hypothetical protein | - |
| SSU05_0919 | 890615..891046 | - | 432 | YP_001198285 | hypothetical protein | - |
| SSU05_0920 | 891541..891894 | + | 354 | YP_001198286 | Tn916, transcriptional regulator | - |
| SSU05_0922 | 892240..894174 | - | 1935 | YP_001198287 | translation elongation factor (GTPases) | - |
| SSU05_0921 | 893067..893219 | + | 153 | YP_001198288 | hypothetical protein | - |
| SSU05_0923 | 894536..895468 | - | 933 | YP_001198289 | Tn916 hypothetical protein | orf13 |
| SSU05_0924 | 895465..896466 | - | 1002 | YP_001198290 | hypothetical protein | orf14 |
| SSU05_0925 | 896463..898640 | - | 2178 | YP_001198291 | hypothetical protein | orf15 |
| SSU05_0926 | 898643..901099 | - | 2457 | YP_001198292 | hypothetical protein | virb4 |
| SSU05_0927 | 901074..901580 | - | 507 | YP_001198293 | hypothetical protein | orf17a |
| SSU05_0928 | 901555..902052 | - | 498 | YP_001198294 | hypothetical protein | - |
| SSU05_0929 | 902169..902390 | - | 222 | YP_001198295 | hypothetical protein | orf19 |
| SSU05_0930 | 902433..903638 | - | 1206 | YP_001198296 | Tn916, transcriptional regulator | - |
| SSU05_0931 | 903816..905201 | - | 1386 | YP_001198297 | hypothetical protein | virb4 |
| SSU05_0932 | 905230..905616 | - | 387 | YP_001198298 | hypothetical protein | orf23 |
| SSU05_0933 | 905632..905946 | - | 315 | YP_001198299 | hypothetical protein | orf23 |
| SSU05_0934 | 907006..908550 | - | 1545 | YP_001198300 | DNA helicase | - |
| SSU05_0935 | 908507..910594 | - | 2088 | YP_001198301 | hypothetical protein | - |
| SSU05_0936 | 910591..911367 | - | 777 | YP_001198302 | Signal recognition particle GTPase | - |
| SSU05_0937 | 911367..911873 | - | 507 | YP_001198303 | transcriptional regulator | - |
| SSU05_0938 | 911913..912266 | - | 354 | YP_001198304 | ATPases with chaperone activity, ATP-binding subunit | - |
| SSU05_0939 | 912253..912642 | - | 390 | YP_001198305 | methyl-accepting chemotaxis protein | - |
| SSU05_0940 | 912639..912866 | - | 228 | YP_001198306 | hypothetical protein | - |
| SSU05_0941 | 912920..913996 | - | 1077 | YP_001198307 | DNA primase (type) | - |
| SSU05_0942 | 914035..914673 | - | 639 | YP_001198308 | hypothetical protein | - |
| SSU05_0943 | 916074..916673 | - | 600 | YP_001198309 | DNA-binding response regulator | - |
| SSU05_0944 | 916651..917841 | - | 1191 | YP_001198310 | two-component sensor histidine kinase | - |
| SSU05_0945 | 918568..918852 | - | 285 | YP_001198311 | hypothetical protein | - |
| SSU05_0946 | 919557..920471 | - | 915 | YP_001198312 | hypothetical protein | - |
| SSU05_0947 | 920443..921546 | - | 1104 | YP_001198313 | multidrug ABC transporter ATPase/permease | - |
| SSU05_0948 | 921488..922372 | - | 885 | YP_001198314 | cytolysin B transport protein | - |
| SSU05_0949 | 922525..923637 | - | 1113 | YP_001198315 | lantibiotic modifying enzyme | - |
| SSU05_0950 | 923737..923856 | + | 120 | YP_001198316 | hypothetical protein | - |
| SSU05_0951 | 923964..924461 | + | 498 | YP_001198317 | DNA recombinase | - |
| SSU05_0952 | 924462..924878 | + | 417 | YP_001198318 | recombinase | - |
| SSU05_0953 | 924880..926442 | + | 1563 | YP_001198319 | DNA recombinase | - |
| SSU05_0954 | 926585..926809 | + | 225 | YP_001198320 | hypothetical protein | - |
| SSU05_0955 | 926824..927693 | + | 870 | YP_001198321 | hypothetical protein | - |
| SSU05_0956 | 927674..928408 | + | 735 | YP_001198322 | hypothetical protein | - |
| SSU05_0957 | 928441..929304 | + | 864 | YP_001198323 | aminoglycoside 6-adenylyltansferase | - |
| SSU05_0958 | 929348..929875 | + | 528 | YP_001198324 | adenine phosphoribosyltransferase | - |
| SSU05_0959 | 930103..931422 | + | 1320 | YP_001198325 | transposase | - |
| SSU05_0960 | 931611..933008 | - | 1398 | YP_001198326 | SalB | - |
| SSU05_0961 | 933916..934215 | - | 300 | YP_001198327 | hypothetical protein | - |
| SSU05_0962 | 934286..941110 | - | 6825 | YP_001198328 | SNF2 family protein | - |
| SSU05_0963 | 941160..941714 | - | 555 | YP_001198329 | hypothetical protein | gbs1354 |
| SSU05_0964 | 941695..941886 | - | 192 | YP_001198330 | hypothetical protein | - |
| SSU05_0965 | 941887..946839 | - | 4953 | YP_001198331 | agglutinin receptor | prgB |
| SSU05_0966 | 946954..947544 | + | 591 | YP_001198332 | transcriptional regulator | - |
| SSU05_0967 | 947541..948386 | + | 846 | YP_001198333 | hypothetical protein | - |
| SSU05_0968 | 948449..951250 | - | 2802 | YP_001198334 | Tn5252, Orf28 | prgK |
| SSU05_0969 | 951252..953603 | - | 2352 | YP_001198335 | Type IV secretory pathway, VirB4 component | virb4 |
| SSU05_0970 | 953560..954015 | - | 456 | YP_001198336 | hypothetical protein | prgIc |
| SSU05_0971 | 953975..954829 | - | 855 | YP_001198337 | cobalt ABC transporter permease CbiQ and related transporters | prgHb |
| SSU05_0972 | 954848..955069 | - | 222 | YP_001198338 | hypothetical protein | prgF |
| SSU05_0973 | 955109..956926 | - | 1818 | YP_001198339 | Type IV secretory pathway, VirD4 component | virb4 |
| SSU05_0974 | 956926..957495 | - | 570 | YP_001198340 | hypothetical protein | gbs1365 |
| SSU05_0975 | 957492..958082 | - | 591 | YP_001198341 | protease | - |
| SSU05_0976 | 958079..958318 | - | 240 | YP_001198342 | hypothetical protein | - |
| SSU05_0977 | 958321..958710 | - | 390 | YP_001198343 | arsenate reductase | - |
| SSU05_0978 | 958715..959143 | - | 429 | YP_001198344 | hypothetical protein | gbs1369 |
| SSU05_0979 | 959127..960482 | - | 1356 | YP_001198345 | C-5 cytosine-specific DNA methylase | - |
| SSU05_0980 | 960534..960638 | - | 105 | YP_001198346 | hypothetical protein | - |
| SSU05_0981 | 960631..961449 | - | 819 | YP_001198347 | hypothetical protein | - |
| SSU05_0982 | 961446..961631 | - | 186 | YP_001198348 | hypothetical protein | - |
| SSU05_0983 | 961818..962183 | - | 366 | YP_001198349 | 50S ribosomal protein L7/L12 | - |
| SSU05_0984 | 962249..962779 | - | 531 | YP_001198350 | ribosomal protein L10 | - |
Host bacterium
| ID | 299 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICE_SsuZYH33_rplL | GenBank | NC_009442 |
| Element size | 2096309 bp | Coordinate of oriT [Strand] | 30698..30830 [-] |
| Host bacterium | Streptococcus suis 05ZYH33 | Coordinate of element | 872952..961832 |
Cargo genes
| Drug resistance gene | tet(M), ant(6)-Ia |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21, AcrIIA8 |