Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200328 |
| Name | oriT_ICE_SsalLAB813_Tn916 |
| Organism | Streptococcus salivarius strain LAB813 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP040804 (2526..2658 [+], 133 nt) |
| oriT length | 133 nt |
| IRs (inverted repeats) | 6..12, 23..29 (ACCCCCC..GGGGGGT) |
| Location of nic site | 75..76 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 133 nt
>oriT_ICE_SsalLAB813_Tn916
ACCTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAATAGCAGAAAATTCTTTGGTTACAAGGAGTTTAGAAAATTTCGTGGTATGTCAAATGAGCTTCAATAGTTGACATAC
ACCTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAATAGCAGAAAATTCTTTGGTTACAAGGAGTTTAGAAAATTTCGTGGTATGTCAAATGAGCTTCAATAGTTGACATAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file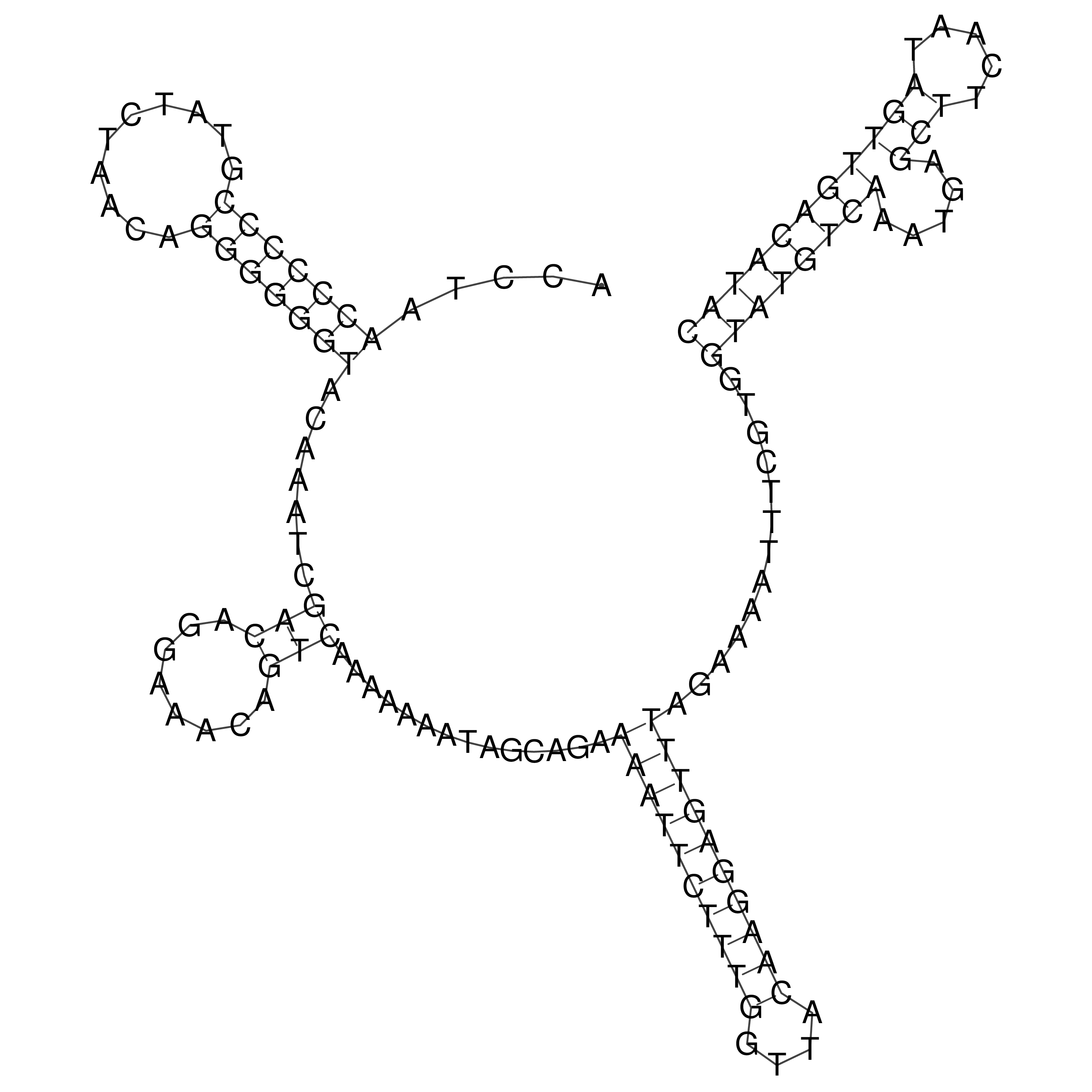
Relaxase
| ID | 14352 | GenBank | QEM31929 |
| Name | Rep_trans_FHI56_03125_ICE_SsalLAB813_Tn916 |
UniProt ID | _ |
| Length | 391 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 391 a.a. Molecular weight: 46169.75 Da Isoelectric Point: 6.5913
>QEM31929.1 XRE family transcriptional regulator [Streptococcus salivarius]
MIQHLKEKRLAYGLSQNRLAIATGITRQYLSDIETGKVKPSDELQQSLWETLERFNPDAPLEMLFDYVRI
RFPTMDVQHVVEEILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHEIDKGVLVELKGRGCRQFESYLL
AQQRSWYEFFMDALVAGGVMKRLDLAINDKTGILNIPTLTEKCLQEECISVFRSFKSYRSGELVRKDEKE
CMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLLVYDNPEHTAF
KIINRYIRFVDKDDSKARSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAPTLKVAITL
DEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MIQHLKEKRLAYGLSQNRLAIATGITRQYLSDIETGKVKPSDELQQSLWETLERFNPDAPLEMLFDYVRI
RFPTMDVQHVVEEILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHEIDKGVLVELKGRGCRQFESYLL
AQQRSWYEFFMDALVAGGVMKRLDLAINDKTGILNIPTLTEKCLQEECISVFRSFKSYRSGELVRKDEKE
CMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLLVYDNPEHTAF
KIINRYIRFVDKDDSKARSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAPTLKVAITL
DEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7875 | GenBank | QEM31944 |
| Name | QEM31944_ICE_SsalLAB813_Tn916 |
UniProt ID | _ |
| Length | 405 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 405 a.a. Molecular weight: 46828.63 Da Isoelectric Point: 9.6601
>QEM31944.1 site-specific integrase [Streptococcus salivarius]
MSEKRRDNKGRILKTGESQRKDGRYLYKYVDSFGEPQFIYSWKLVATDRVPAGKRDCVPLREKVAELLKD
IHDGIDVVGKKMTLCQLYAKQNAQRPNVKKGTQTGRKYLMDILTKDKLGARSIDSIKPSDAKEWAIRMSE
DGYAYQTINNYKRSLRASFYIAIEDDCVRKNPFDFQLNTVIDDDTVPKVALTTEQEEKLLSFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVKVDHQLLRDTENGYYIETPKTKSGERQVPMVEEAYQ
AFQRVLANRKNRKCVVIDGYSNFLFLNGKDYPKVASDYNGMLKNLVKKYNKYHEDKLPHITPHILRHTFC
TNYANAGMNPKALQYIMGHANINMTLNYYAHATFDSAMAEMNRLEKKKQQERLVA
MSEKRRDNKGRILKTGESQRKDGRYLYKYVDSFGEPQFIYSWKLVATDRVPAGKRDCVPLREKVAELLKD
IHDGIDVVGKKMTLCQLYAKQNAQRPNVKKGTQTGRKYLMDILTKDKLGARSIDSIKPSDAKEWAIRMSE
DGYAYQTINNYKRSLRASFYIAIEDDCVRKNPFDFQLNTVIDDDTVPKVALTTEQEEKLLSFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVKVDHQLLRDTENGYYIETPKTKSGERQVPMVEEAYQ
AFQRVLANRKNRKCVVIDGYSNFLFLNGKDYPKVASDYNGMLKNLVKKYNKYHEDKLPHITPHILRHTFC
TNYANAGMNPKALQYIMGHANINMTLNYYAHATFDSAMAEMNRLEKKKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16845 | GenBank | QEM31927 |
| Name | tcpA_FHI56_03115_ICE_SsalLAB813_Tn916 |
UniProt ID | _ |
| Length | 464 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 464 a.a. Molecular weight: 53690.45 Da Isoelectric Point: 8.2463
>QEM31927.1 DNA translocase FtsK [Streptococcus salivarius]
MKQFGIRGKRIRPSDKDLVFHFTVASLLPIFLLVIGLFHVKTIQQINWQDFNLSQVDKIHIPYLVISVSV
AILVCLLVAFLFKRYRYDTVKQLYHRQKLAKMVLENKWYESEQVKTDGFFKDSSSRTKEKITYFPKIFYR
LKDGLIQMRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVQAKDGK
LRLMENVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLYTDSKLYILDPKNADLADLGSVMGNVYYRKE
DMLSCIDRFYDEMMARSEAMKEMENYKTGENYAYLGLPANFLIFDEYVAFMEMLGNKENTAVLNKLKQIV
MLGRQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKQIKGRGYVDVG
TSVISEFYTPLVPKGHDFLEEIKKLTNSRQSTQATCEAKVAGVD
MKQFGIRGKRIRPSDKDLVFHFTVASLLPIFLLVIGLFHVKTIQQINWQDFNLSQVDKIHIPYLVISVSV
AILVCLLVAFLFKRYRYDTVKQLYHRQKLAKMVLENKWYESEQVKTDGFFKDSSSRTKEKITYFPKIFYR
LKDGLIQMRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVQAKDGK
LRLMENVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLYTDSKLYILDPKNADLADLGSVMGNVYYRKE
DMLSCIDRFYDEMMARSEAMKEMENYKTGENYAYLGLPANFLIFDEYVAFMEMLGNKENTAVLNKLKQIV
MLGRQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKQIKGRGYVDVG
TSVISEFYTPLVPKGHDFLEEIKKLTNSRQSTQATCEAKVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 385673..397026
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| FHI56_03085 | 381285..383924 | + | 2640 | QEM31922 | DNA polymerase I | - |
| FHI56_03090 | 384479..384880 | + | 402 | QEM31923 | hypothetical protein | - |
| FHI56_03095 | 384901..385161 | + | 261 | QEM31924 | hypothetical protein | - |
| FHI56_03100 | 385525..385651 | + | 127 | Protein_366 | hypothetical protein | - |
| FHI56_03105 | 385673..385987 | + | 315 | QEM31925 | DUF961 domain-containing protein | orf23 |
| FHI56_03110 | 386003..386389 | + | 387 | QEM31926 | DUF961 domain-containing protein | orf23 |
| FHI56_03115 | 386429..387823 | + | 1395 | QEM31927 | DNA translocase FtsK | virb4 |
| FHI56_03120 | 387826..387981 | + | 156 | QEM31928 | conjugal transfer protein | - |
| FHI56_03125 | 388029..389204 | + | 1176 | QEM31929 | XRE family transcriptional regulator | - |
| FHI56_03130 | 389247..389468 | + | 222 | QEM31930 | hypothetical protein | orf19 |
| FHI56_03135 | 389585..390082 | + | 498 | QEM31931 | antirestriction protein ArdA | - |
| FHI56_03140 | 390096..390488 | + | 393 | QEM31932 | conjugal transfer protein | orf17a |
| FHI56_03145 | 390472..392919 | + | 2448 | QEM31933 | ATP-binding protein | virb4 |
| FHI56_03150 | 392922..395099 | + | 2178 | QEM31934 | YtxH domain-containing protein | orf15 |
| FHI56_03155 | 395096..396097 | + | 1002 | QEM31935 | peptidase P60 | orf14 |
| FHI56_03160 | 396094..397026 | + | 933 | QEM31936 | conjugal transfer protein | orf13 |
| FHI56_03165 | 397280..397387 | + | 108 | QEM31937 | tetracycline resistance determinant leader peptide | - |
| FHI56_03170 | 397403..399322 | + | 1920 | QEM31938 | tetracycline resistance ribosomal protection protein Tet(M) | - |
| FHI56_03175 | 399413..399607 | + | 195 | QEM33470 | conjugal transfer protein | - |
| FHI56_03180 | 399664..400017 | - | 354 | QEM31939 | helix-turn-helix transcriptional regulator | - |
| FHI56_03185 | 400528..400953 | + | 426 | QEM31940 | sigma-70 family RNA polymerase sigma factor | - |
| FHI56_03190 | 400950..401180 | + | 231 | QEM31941 | helix-turn-helix domain-containing protein | - |
| FHI56_03195 | 401411..401548 | - | 138 | QEM31942 | conjugal transfer protein | - |
| FHI56_03200 | 401642..401845 | + | 204 | QEM31943 | excisionase | - |
Host bacterium
| ID | 293 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICE_SsalLAB813_Tn916 | GenBank | CP040804 |
| Element size | 2242557 bp | Coordinate of oriT [Strand] | 2526..2658 [+] |
| Host bacterium | Streptococcus salivarius strain LAB813 | Coordinate of element | 385333..403318 |
Cargo genes
| Drug resistance gene | tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |