Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200305 |
| Name | oriT_ICE_SdyRE378_TnGBS2 |
| Organism | Streptococcus dysgalactiae subsp. equisimilis RE378 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_018712 (5943..5975 [-], 33 nt) |
| oriT length | 33 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 33 nt
>oriT_ICE_SdyRE378_TnGBS2
GCATGTGTCCGGACACGTGCTACGCTTGCCAGA
GCATGTGTCCGGACACGTGCTACGCTTGCCAGA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file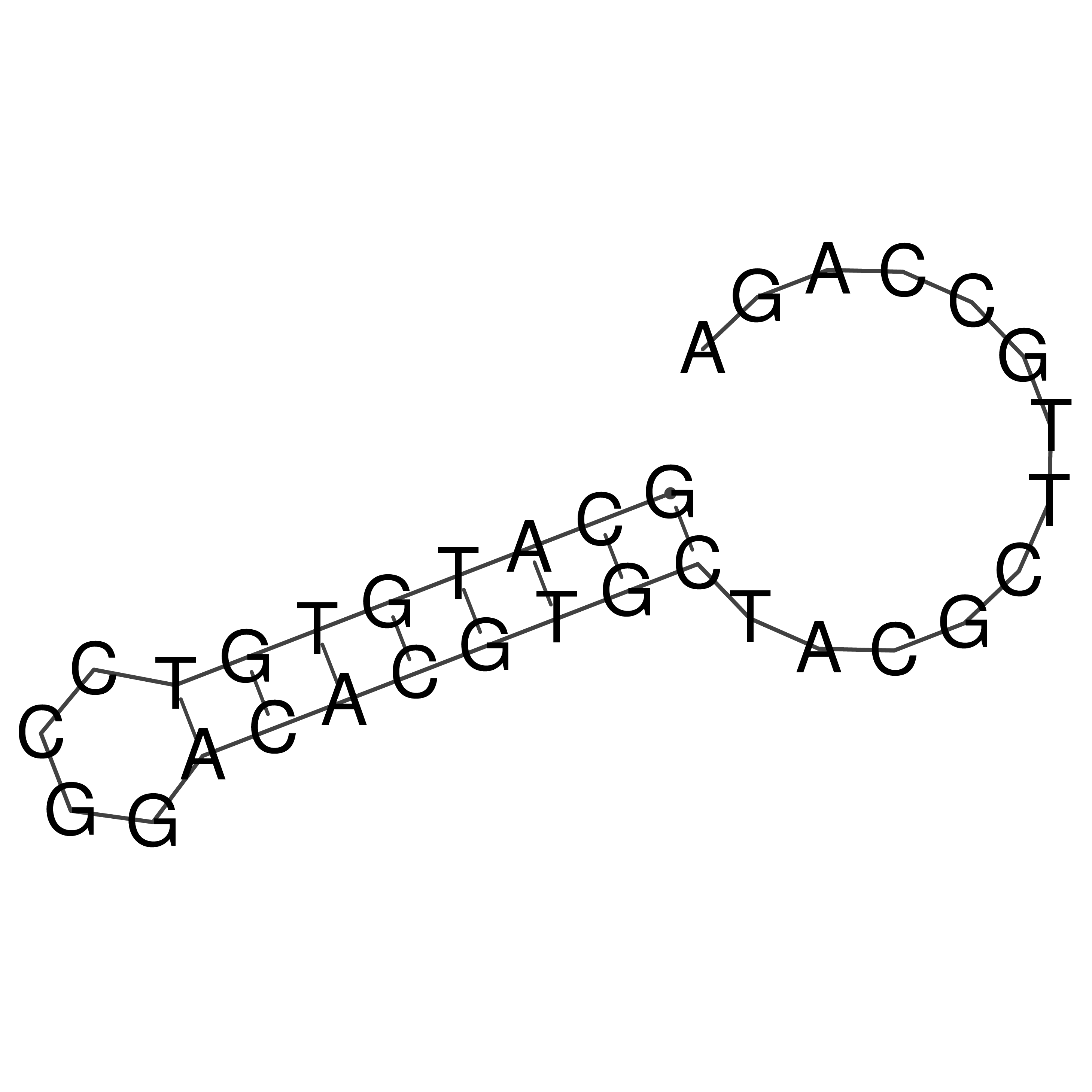
Relaxase
| ID | 14345 | GenBank | WP_000261836 |
| Name | Relaxase_GGS_RS07840_ICE_SdyRE378_TnGBS2 |
UniProt ID | Q8E5B1 |
| Length | 543 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 543 a.a. Molecular weight: 63241.72 Da Isoelectric Point: 6.9556
>WP_000261836.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Streptococcus]
MVVTKVIQIKSSKNLKRAINYIARDDATLIKGNGNLADEDYSYEVVNGQVMKRLVSGHDLTDISDTDMIY
EDFMLLKQSVDALYDNDTLSDLKNPNRVLAHHVIQSFSPNDELTPEEVNEIGRKTALELTGGDYQFVVAT
HQDKGHLHNHIIFSTTNQVTLKKFRWQKSTKKSLEHISDKHAELYGAKILEPKLRNSYTDYSAWRRKNNF
RYEIKERLDFLLKHSLDKDDFFQKAKALNLHIDTSGKYVTYKLIDSPQERPVRDRTLSKKGKYYLDKMVE
RFAINEVVYNLDHIKEKYDEEQAKKAEDFEMKVRIEPWQIKQLTNQSIHVSIIFGLDRQGTVAIPARMLD
QNEDGSFTAYLKKNDFFYFLNADHSEQNRFVKGTTLIKQLSAQNGEVILTKNKHVAELNRLVDEFNFLAA
NQVTDSTQFMQLKETFLEQLVETDKTLEQLDDKMTYLNKVLGALMDYQNGIIPSEVTMTILDKAKVDKDG
DTKSLRKEIKELQIERETLHKVRDQIVNDYDFAIEMTNRYDKSNKESKRRSML
MVVTKVIQIKSSKNLKRAINYIARDDATLIKGNGNLADEDYSYEVVNGQVMKRLVSGHDLTDISDTDMIY
EDFMLLKQSVDALYDNDTLSDLKNPNRVLAHHVIQSFSPNDELTPEEVNEIGRKTALELTGGDYQFVVAT
HQDKGHLHNHIIFSTTNQVTLKKFRWQKSTKKSLEHISDKHAELYGAKILEPKLRNSYTDYSAWRRKNNF
RYEIKERLDFLLKHSLDKDDFFQKAKALNLHIDTSGKYVTYKLIDSPQERPVRDRTLSKKGKYYLDKMVE
RFAINEVVYNLDHIKEKYDEEQAKKAEDFEMKVRIEPWQIKQLTNQSIHVSIIFGLDRQGTVAIPARMLD
QNEDGSFTAYLKKNDFFYFLNADHSEQNRFVKGTTLIKQLSAQNGEVILTKNKHVAELNRLVDEFNFLAA
NQVTDSTQFMQLKETFLEQLVETDKTLEQLDDKMTYLNKVLGALMDYQNGIIPSEVTMTILDKAKVDKDG
DTKSLRKEIKELQIERETLHKVRDQIVNDYDFAIEMTNRYDKSNKESKRRSML
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q8E5B1 |
T4CP
| ID | 16837 | GenBank | WP_000421159 |
| Name | t4cp2_GGS_RS07870_ICE_SdyRE378_TnGBS2 |
UniProt ID | _ |
| Length | 681 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 681 a.a. Molecular weight: 78354.02 Da Isoelectric Point: 5.1508
>WP_000421159.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Streptococcus]
MELKRKRLLPYLLLSLLLAYLTHRAYVLYCMAPQADMTNLFAPYTYVFEKITEPPYFYWNTSPLAILSAL
VGFFVGLLFYLKIRLDGNYRHGEESGSACFATAQELKGFQDEESRNNMIFSREAQMGLFNKRLPFERQLN
KNVLVAGLPGDGKTYTYVKPNLMQMNSSFIVTDPKGLLVREVGTMLKENGYQIKIFDLVNLTNSDMFNPF
RYMRSELDIDRITEAIVEGTKKGDREGEDFWNQAKLLLNRALLGYLYFDSQVRHYEPNLSMVADLLRNLK
RPNEKELSAVEKMFKQLEEKLPGNYACRQWELFNSNFEAETRTGVLSIVATQYSIFDHDAVTNLIKTDTM
EMDTWNTEKTAVFVAISETNKAYSFLASTFFTVVFDQLTHSADAIIQGEKKGYELKDLLHVQFYFDEFAN
CGKIPHFNEVLASIRSREMSIKIIIQAISQLDSLYGVPARKSMVNNCATLLFLGTNDEDTMKYFSMRAGK
QTIIQTSYSEQRGHRISGTTSRQAHQRDLMTPDEIARIGVDEALVFISKQNVFRDKKAMVSNHPMKDRLA
NHYQDKTWYHYKRFMEEGAEFIDAVMTGQIPEDRIWAPDMSDYQAFLEENGFDNTRQSAPETSVHNQVPV
TNPEMPSESPSQTSETSKKLVDMDTGEIFELPPEDREGFGEVSFDDQRVEI
MELKRKRLLPYLLLSLLLAYLTHRAYVLYCMAPQADMTNLFAPYTYVFEKITEPPYFYWNTSPLAILSAL
VGFFVGLLFYLKIRLDGNYRHGEESGSACFATAQELKGFQDEESRNNMIFSREAQMGLFNKRLPFERQLN
KNVLVAGLPGDGKTYTYVKPNLMQMNSSFIVTDPKGLLVREVGTMLKENGYQIKIFDLVNLTNSDMFNPF
RYMRSELDIDRITEAIVEGTKKGDREGEDFWNQAKLLLNRALLGYLYFDSQVRHYEPNLSMVADLLRNLK
RPNEKELSAVEKMFKQLEEKLPGNYACRQWELFNSNFEAETRTGVLSIVATQYSIFDHDAVTNLIKTDTM
EMDTWNTEKTAVFVAISETNKAYSFLASTFFTVVFDQLTHSADAIIQGEKKGYELKDLLHVQFYFDEFAN
CGKIPHFNEVLASIRSREMSIKIIIQAISQLDSLYGVPARKSMVNNCATLLFLGTNDEDTMKYFSMRAGK
QTIIQTSYSEQRGHRISGTTSRQAHQRDLMTPDEIARIGVDEALVFISKQNVFRDKKAMVSNHPMKDRLA
NHYQDKTWYHYKRFMEEGAEFIDAVMTGQIPEDRIWAPDMSDYQAFLEENGFDNTRQSAPETSVHNQVPV
TNPEMPSESPSQTSETSKKLVDMDTGEIFELPPEDREGFGEVSFDDQRVEI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1614901..1630074
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GGS_RS07865 (GGS_1495) | 1612449..1612814 | - | 366 | WP_000188894 | hypothetical protein | - |
| GGS_RS07870 (GGS_1496) | 1612856..1614901 | - | 2046 | WP_000421159 | VirD4-like conjugal transfer protein, CD1115 family | - |
| GGS_RS07875 (GGS_1497) | 1614901..1615392 | - | 492 | WP_000371901 | DUF3801 domain-containing protein | cd411 |
| GGS_RS07880 (GGS_1498) | 1615414..1616196 | - | 783 | WP_000488485 | hypothetical protein | - |
| GGS_RS07885 | 1616196..1616468 | - | 273 | WP_000606554 | hypothetical protein | - |
| GGS_RS07890 (GGS_1499) | 1616488..1617093 | - | 606 | WP_000567358 | hypothetical protein | prgL |
| GGS_RS07895 (GGS_1500) | 1617114..1619804 | - | 2691 | WP_000134095 | phage tail tip lysozyme | prgK |
| GGS_RS10965 | 1620293..1620436 | - | 144 | WP_017645960 | hypothetical protein | - |
| GGS_RS07905 (GGS_1502) | 1620441..1622792 | - | 2352 | WP_000176428 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| GGS_RS07910 (GGS_1503) | 1622773..1623132 | - | 360 | WP_001037292 | PrgI family protein | - |
| GGS_RS07915 (GGS_1504) | 1623194..1623652 | - | 459 | WP_000609828 | hypothetical protein | - |
| GGS_RS07920 | 1623713..1623940 | - | 228 | WP_000362731 | hypothetical protein | - |
| GGS_RS11215 | 1624184..1624492 | - | 309 | WP_231846834 | hypothetical protein | - |
| GGS_RS07930 (GGS_1506) | 1624695..1625447 | - | 753 | WP_000651825 | membrane protein | prgHb |
| GGS_RS07935 (GGS_1507) | 1625496..1625981 | - | 486 | WP_000004254 | hypothetical protein | - |
| GGS_RS07940 | 1626087..1626314 | - | 228 | WP_000447008 | hypothetical protein | prgF |
| GGS_RS07945 (GGS_1508) | 1626589..1629387 | - | 2799 | WP_001088030 | SspB-related isopeptide-forming adhesin | prgB |
| GGS_RS07950 (GGS_1509) | 1629454..1630074 | - | 621 | WP_015017288 | LPXTG cell wall anchor domain-containing protein | prgC |
| GGS_RS07955 (GGS_1510) | 1630089..1632320 | - | 2232 | WP_001071823 | LPXTG cell wall anchor domain-containing protein | - |
| GGS_RS07960 (GGS_1511) | 1632337..1632636 | - | 300 | WP_000129105 | hypothetical protein | - |
| GGS_RS07965 | 1633019..1633216 | - | 198 | WP_000208122 | helix-turn-helix transcriptional regulator | - |
| GGS_RS07970 | 1633213..1633398 | - | 186 | WP_000938679 | hypothetical protein | - |
| GGS_RS07975 (GGS_1512) | 1633400..1634425 | - | 1026 | WP_000822468 | replication initiator protein A | - |
| GGS_RS11035 | 1634427..1634591 | - | 165 | WP_000166480 | hypothetical protein | - |
Host bacterium
| ID | 273 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICE_SdyRE378_TnGBS2 | GenBank | NC_018712 |
| Element size | 2151145 bp | Coordinate of oriT [Strand] | 5943..5975 [-] |
| Host bacterium | Streptococcus dysgalactiae subsp. equisimilis RE378 | Coordinate of element | 1602651..1636331 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7, AcrIIA21 |