Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200304 |
| Name | oriT_ICE_FprA2-165_rumA |
| Organism | Faecalibacterium duncaniae strain JCM 31915 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP048437 (33130..33157 [+], 28 nt) |
| oriT length | 28 nt |
| IRs (inverted repeats) | 1..7, 12..18 (GGGAGTG..CACTCCC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 28 nt
GGGAGTGGCTACACTCCCTATGCTTGCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file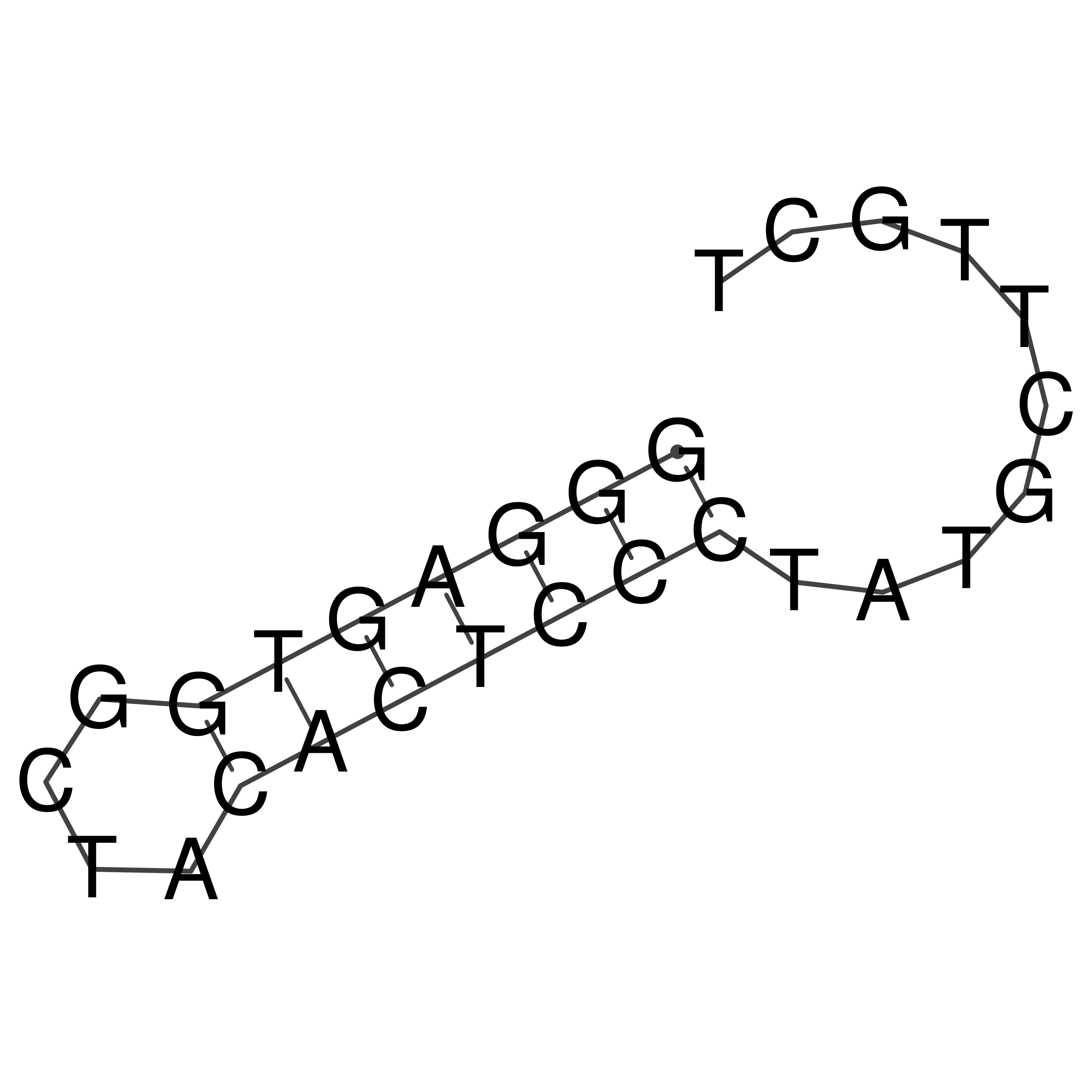
Relaxase
| ID | 14342 | GenBank | QIA43153 |
| Name | Relaxase_GXM22_08770_ICE_FprA2-165_rumA |
UniProt ID | _ |
| Length | 442 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 442 a.a. Molecular weight: 50761.82 Da Isoelectric Point: 9.4661
MAVTKIHPIKSTLKKALDYIENPVKTDEKILVSSFACSYETADIEFELLLSQAMQKGNNLAHHLIQSFAP
GETTPEQAHEIGRQLADEVLQGKYPYVLTTHIDKGHVHNHIIFCAVDLVNQRKYVSNRQSYSYIRRTSDR
LCKEHNLSVVMPGQDRGKSYAEWNAHRKGTSWKAKLKAAIDTTIPQAKDFDDFLRLLQEQSYEVKRGKYV
SFRAPGQERFTRCKTLGEAYTEEAITERIKGRFAERKPKETRKISLQIDLENSIKAQQSAGYEKWAKLHN
LKQAARTLNFLTEHEIDSYPDLESRVAEITAVSTEAAAALKTAERRLAEMAVLIKDVTTCKELRPLVQEY
QQAADKKQFRRKHEGTLILYEAAAKALKEQGFQKLPDLYALKAEYKQLAEQKDQLQRQYNDAKRQMQEYG
IIKQNVDGILRTAPGKEQMQER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 14343 | GenBank | QIA44352 |
| Name | MobA_MobL_GXM22_08825_ICE_FprA2-165_rumA |
UniProt ID | _ |
| Length | 471 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 471 a.a. Molecular weight: 54978.44 Da Isoelectric Point: 8.5444
MASAAYRAGERLYSSYYGEVSDYTKKGGVIASEIMLPPHAPEIYLDRATLWNAVENCEKHPKAQLAYSFD
IAMQNELTLEENMELARKFVQEQFVTKGMIADLAFHSPEKEDGGIPNPHFHVMTTMRPLNPDGTWGQKQR
REYLLDEDGNRIRDKNGDYMFNAVHTTDWHEPETLEHWREQWAAAVNTKFEEKGLDVRIDHRSYVRQGLD
LIPTVHEGANVRQMEAKGIRTEKGELNRWIKATNRLMQDVRKKIKALFVWMAEVKEELSKPQTPSLADLL
IAYYNQRNAGAWSNKARTGNLKQFAEAVNYLTENKLLTLEDLQERLSSVNEEFEALSDSMKKKSARIKEL
QELIREGENYQRLKPVHTELNNIKFKKQREKFETSHDAELRLFYAARRILKEKLDGKPIALKAWKQEYAQ
LKTEYAELSPQHKPLREEVIRLRQVQNAVDTALRRREQPQAVQRKKHEMEL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7872 | GenBank | QIA43154 |
| Name | QIA43154_ICE_FprA2-165_rumA |
UniProt ID | _ |
| Length | 109 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 109 a.a. Molecular weight: 12619.71 Da Isoelectric Point: 11.0031
MTNRTRKIVLRVPVTPEERELIRQKMALLHTRNFSAYARKMLIDGYVVHIDTTGIREQTAELQKIGVNVN
QIARRLNSMGPLYTQDVADIKGALAQIWQLQRYILSSQR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16835 | GenBank | QIA43128 |
| Name | t4cp2_GXM22_08620_ICE_FprA2-165_rumA |
UniProt ID | _ |
| Length | 601 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 601 a.a. Molecular weight: 67732.63 Da Isoelectric Point: 6.7002
MKPELKKLLVLNLPYLLFVYLFAKCGQAYRLAAGADASAKLLHLTGGISVAFASPLPSLHLFDLCVGVVG
AVAVRLIVYSKGKNAKKYRKGEEYGSARWGTAKDIAPYIDPKFENNILLTQTERLTMTGRPKDPKTARNK
NVLVIGGSGSGKTRFYVKPNLMQCFPTSDYPTSFVVTDPKGTLVLETGQMFQRAGYRVKILNTINFSKSM
KYNPFVYIHSEKDVLKLVNTLIANTKGEGEKSAEDFWVKSERLFYTALIGYIWYEAPAEEMNFTTLLEMI
NASEAREDDPEFQSPVDLMFERLEQKDPDHFAVRQYKKFLLSAGKTRSSILISCGARLAPFDIREVRELM
EDDEMELDTIGDEKTVLFLIMSDTDTTFNFILAMLQSQLINLLCDRADDKYGGRLPVHVRLILDEFANIG
QIPNFDKLIATIRSREISASIILQSQSQLKAIYKDAAEIISDNCDSVLFLSGRGKNAKEISDALGKETID
SFNTSENRGSQTSHGLNYQKLGKALMSEDEIAIMDGGRCILQLRGVRPFFSEKFDITKHPHYKYLADADK
KNTFDVDRFLSSLRRKRQQVVAQDESFDLYEIDLSDENAAE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1762683..1784231
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GXM22_08600 | 1759175..1760038 | + | 864 | QIA43124 | AraC family transcriptional regulator | - |
| GXM22_08605 | 1760067..1761440 | + | 1374 | QIA43125 | 23S rRNA (uracil(1939)-C(5))-methyltransferase RlmD | - |
| GXM22_08610 | 1761668..1762582 | + | 915 | QIA43126 | helix-turn-helix domain-containing protein | - |
| GXM22_08615 | 1762683..1763153 | + | 471 | QIA43127 | PcfB family protein | cd411 |
| GXM22_08620 | 1763150..1764955 | + | 1806 | QIA43128 | type IV secretory system conjugative DNA transfer family protein | - |
| GXM22_08625 | 1765003..1765143 | + | 141 | QIA43129 | hypothetical protein | prgF |
| GXM22_08630 | 1765348..1765725 | + | 378 | QIA43130 | GntR family transcriptional regulator | - |
| GXM22_08635 | 1765728..1766588 | + | 861 | QIA43131 | ABC transporter ATP-binding protein | - |
| GXM22_08640 | 1766585..1767232 | + | 648 | QIA43132 | ABC-2 transporter permease | - |
| GXM22_08645 | 1767573..1768016 | + | 444 | QIA43133 | sigma-70 family RNA polymerase sigma factor | - |
| GXM22_08650 | 1768009..1768209 | + | 201 | QIA43134 | helix-turn-helix domain-containing protein | - |
| GXM22_08655 | 1768676..1769101 | + | 426 | QIA44349 | plasmid mobilization relaxosome protein MobC | - |
| GXM22_08660 | 1769160..1770693 | + | 1534 | Protein_1688 | relaxase/mobilization nuclease domain-containing protein | - |
| GXM22_08665 | 1770867..1771061 | + | 195 | QIA44350 | hypothetical protein | - |
| GXM22_08670 | 1771051..1771791 | + | 741 | QIA43135 | replication protein | - |
| GXM22_08675 | 1771788..1772627 | + | 840 | QIA43136 | ATP-binding protein | - |
| GXM22_08680 | 1772640..1772831 | + | 192 | QIA43137 | hypothetical protein | - |
| GXM22_08685 | 1772966..1774573 | + | 1608 | QIA43138 | DUF4368 domain-containing protein | - |
| GXM22_08690 | 1774592..1774756 | + | 165 | Protein_1694 | hypothetical protein | - |
| GXM22_08695 | 1774860..1775525 | + | 666 | QIA43139 | hypothetical protein | prgHa |
| GXM22_08700 | 1776101..1777933 | + | 1833 | QIA43140 | group II intron reverse transcriptase/maturase | - |
| GXM22_08705 | 1778009..1778311 | + | 303 | QIA43141 | hypothetical protein | prgHa |
| GXM22_08710 | 1778332..1778757 | + | 426 | QIA43142 | PrgI family protein | prgIa |
| GXM22_08715 | 1778684..1781086 | + | 2403 | QIA43143 | PrgI family protein | virb4 |
| GXM22_08720 | 1781083..1781979 | + | 897 | QIA43144 | DNA (cytosine-5-)-methyltransferase | - |
| GXM22_08725 | 1781976..1783961 | + | 1986 | QIA43145 | C40 family peptidase | cd419a |
| GXM22_08730 | 1783980..1784231 | + | 252 | QIA43146 | DUF4315 family protein | cd419 |
| GXM22_08735 | 1784221..1784943 | + | 723 | QIA43147 | DUF4366 domain-containing protein | - |
| GXM22_08740 | 1784940..1787063 | + | 2124 | QIA43148 | DNA topoisomerase III | - |
Region 2: 2998030..3009698
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GXM22_14455 | 2994438..2995241 | - | 804 | QIA44164 | phosphoadenosine phosphosulfate reductase | - |
| GXM22_14460 | 2995192..2997261 | - | 2070 | QIA44165 | DNA topoisomerase III | - |
| GXM22_14465 | 2997258..2998037 | - | 780 | QIA44166 | DUF4366 domain-containing protein | - |
| GXM22_14470 | 2998030..2998266 | - | 237 | QIA44167 | DUF4315 family protein | cd419 |
| GXM22_14475 | 2998283..3000490 | - | 2208 | QIA44168 | peptidase M23 | cd419a |
| GXM22_14480 | 3000490..3001443 | - | 954 | QIA44410 | site-specific DNA-methyltransferase | - |
| GXM22_14485 | 3001472..3002047 | - | 576 | QIA44169 | nitrogen fixation protein | - |
| GXM22_14490 | 3002063..3004462 | - | 2400 | QIA44411 | conjugal transfer protein TraE | virb4 |
| GXM22_14495 | 3004464..3004844 | - | 381 | QIA44170 | PrgI family protein | prgIa |
| GXM22_14500 | 3004813..3005301 | - | 489 | QIA44171 | hypothetical protein | - |
| GXM22_14505 | 3005289..3005882 | - | 594 | QIA44172 | adenine methyltransferase | - |
| GXM22_14510 | 3005872..3006741 | - | 870 | QIA44173 | hypothetical protein | prgHa |
| GXM22_14515 | 3006781..3006996 | - | 216 | QIA44174 | conjugal transfer protein | prgF |
| GXM22_14520 | 3007091..3007306 | - | 216 | QIA44175 | conjugal transfer protein | prgF |
| GXM22_14525 | 3007441..3009210 | - | 1770 | QIA44176 | type IV secretory system conjugative DNA transfer family protein | virb4 |
| GXM22_14530 | 3009207..3009698 | - | 492 | QIA44177 | PcfB family protein | cd411 |
| GXM22_14535 | 3009829..3010734 | - | 906 | QIA44178 | helix-turn-helix domain-containing protein | - |
| GXM22_14540 | 3010731..3011504 | - | 774 | QIA44179 | toxin Bro | - |
| GXM22_14545 | 3011497..3011745 | - | 249 | QIA44180 | hypothetical protein | - |
| GXM22_14550 | 3011872..3012180 | - | 309 | QIA44181 | thiamine transporter | - |
| GXM22_14555 | 3012334..3012549 | - | 216 | Protein_2833 | recombinase family protein | - |
| GXM22_14560 | 3012695..3012889 | - | 195 | QIA44182 | hypothetical protein | - |
| GXM22_14565 | 3012962..3013906 | - | 945 | QIA44183 | plasmid recombination protein | - |
| GXM22_14570 | 3013878..3014065 | - | 188 | Protein_2836 | hypothetical protein | - |
| GXM22_14575 | 3014090..3014485 | - | 396 | QIA44184 | replication initiator protein A | - |
Host bacterium
| ID | 272 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICE_FprA2-165_rumA | GenBank | CP048437 |
| Element size | 3102523 bp | Coordinate of oriT [Strand] | 33130..33157 [+] |
| Host bacterium | Faecalibacterium duncaniae strain JCM 31915 | Coordinate of element | 1761411..1808878 |
Cargo genes
| Drug resistance gene | ant(6)-Ia |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21, AcrIIA9 |