Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200261 |
| Name | oriT_ICEMloMAF-1 |
| Organism | Mesorhizobium loti MAFF303099 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | BA000012 (367747..367804 [-], 58 nt) |
| oriT length | 58 nt |
| IRs (inverted repeats) | 27..33, 37..43 (CCGCCTC..GAGGCGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 58 nt
>oriT_ICEMloMAF-1
AAAAGGATGTGCAGACAAAGGCTTCGCCGCCTCATGGAGGCGGTGCCTTTATCTTGCC
AAAAGGATGTGCAGACAAAGGCTTCGCCGCCTCATGGAGGCGGTGCCTTTATCTTGCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file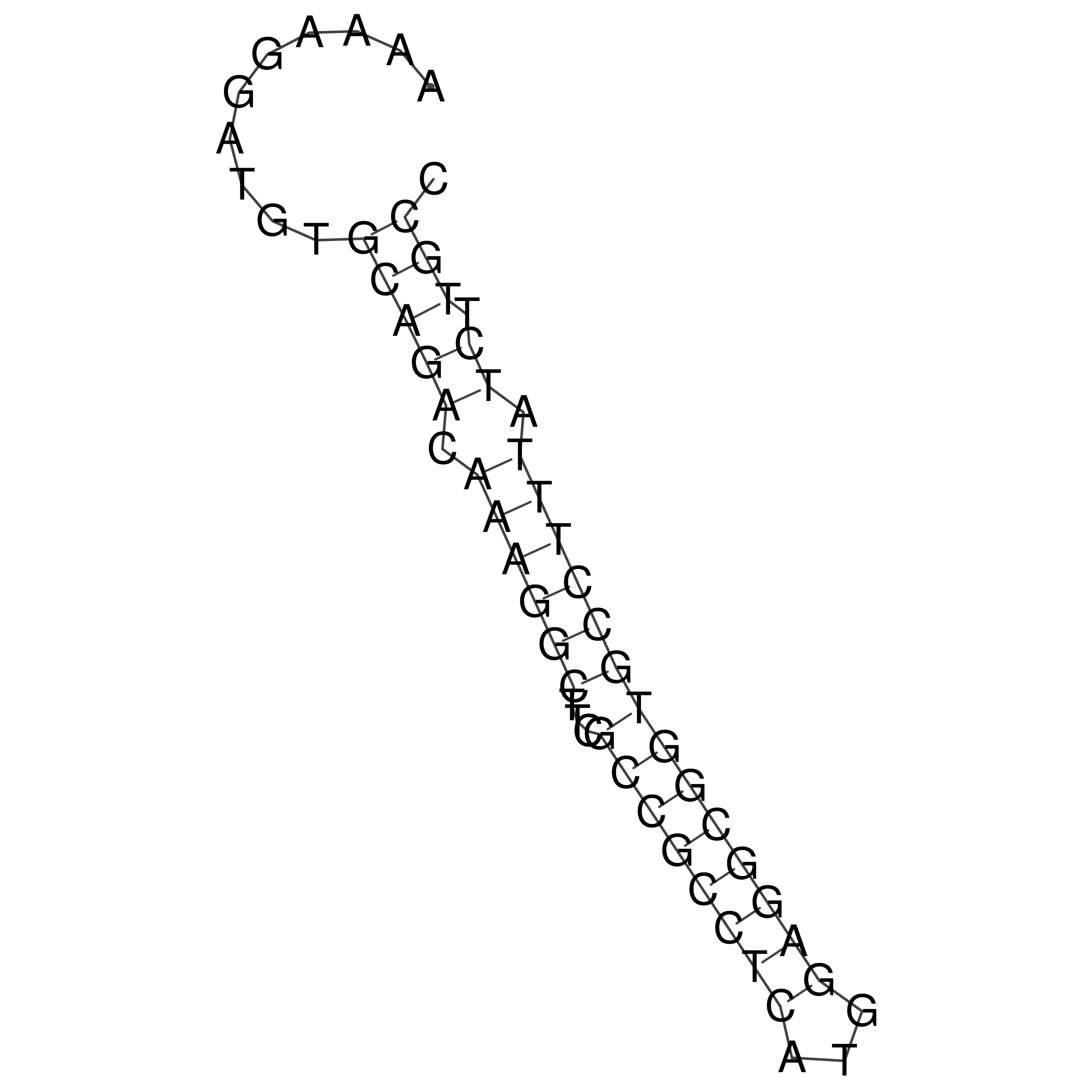
T4CP
| ID | 16768 | GenBank | BAB52699 |
| Name | mlr6395_-_ICEMloMAF-1 |
UniProt ID | _ |
| Length | 735 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 735 a.a. Molecular weight: 81570.91 Da Isoelectric Point: 7.7957
>BAB52699.1 conjugal transfer protein; TraG [Mesorhizobium japonicum MAFF 303099]
MSPLRHSSPRECANIDTPRKRATARQTAPHRTKSYIGRIPVPAVPGDARSISPEQRSKPVTPTKLLIGQI
AIVFAIVLLGVWAATQWCAHMLAFQEQLGAPWFVAAGWPIYEPWKLLEWWFQFDAYAPEVFDKAGMLAGT
SGFMGCAAAIAGSLWRARQRGLVTTYGSSRWAATREIEKAGLFQPAGVFLGKLKDRYLRHDGPEHVMAFA
PTRSGKGVGLVVPTLLSWSGSAVIHDIKGENWQLSSGWRSRFSHCLLFNPTDPRSARYNPLLEVRKGSDE
IRDVQNIADILVDPEGALERRNHWEKTSHSLLVGAILHVLYAEEEKTLARVATFLSDPQRSFAATLRRMM
TTNHLGAANKPQVHPVVASAAREVLNKSENERSGVLSTAMSFLGLYRDPTVAAVTSDCDWRIADLMDAEW
PMSLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPKKSRKHQLLMMLDEFPALGRLDFFETALAFM
AGYGIRAYLIAQSLNQISKAYGDNNAILDNCHLRIAFSSNDERTAKRISDALGTATELRSMRNYAGHRLA
PWLSHVMVSRQETARPLLTPGEVMQLPPADELVLVSGLPPIRAKKLRYYEDHNFTERVLPSPVLRDGPYA
DCPMSRPDDWTGQVRGVDRRLASDDESAGAALADEGGVQQQRHPGLPEEHTAVTQEPDQDDLSRPADDDS
EAMADKRVMDRISTVAGAYGINEGRGDDKDIVPGF
MSPLRHSSPRECANIDTPRKRATARQTAPHRTKSYIGRIPVPAVPGDARSISPEQRSKPVTPTKLLIGQI
AIVFAIVLLGVWAATQWCAHMLAFQEQLGAPWFVAAGWPIYEPWKLLEWWFQFDAYAPEVFDKAGMLAGT
SGFMGCAAAIAGSLWRARQRGLVTTYGSSRWAATREIEKAGLFQPAGVFLGKLKDRYLRHDGPEHVMAFA
PTRSGKGVGLVVPTLLSWSGSAVIHDIKGENWQLSSGWRSRFSHCLLFNPTDPRSARYNPLLEVRKGSDE
IRDVQNIADILVDPEGALERRNHWEKTSHSLLVGAILHVLYAEEEKTLARVATFLSDPQRSFAATLRRMM
TTNHLGAANKPQVHPVVASAAREVLNKSENERSGVLSTAMSFLGLYRDPTVAAVTSDCDWRIADLMDAEW
PMSLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPKKSRKHQLLMMLDEFPALGRLDFFETALAFM
AGYGIRAYLIAQSLNQISKAYGDNNAILDNCHLRIAFSSNDERTAKRISDALGTATELRSMRNYAGHRLA
PWLSHVMVSRQETARPLLTPGEVMQLPPADELVLVSGLPPIRAKKLRYYEDHNFTERVLPSPVLRDGPYA
DCPMSRPDDWTGQVRGVDRRLASDDESAGAALADEGGVQQQRHPGLPEEHTAVTQEPDQDDLSRPADDDS
EAMADKRVMDRISTVAGAYGINEGRGDDKDIVPGF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16769 | GenBank | BAB52704 |
| Name | mlr6400_-_ICEMloMAF-1 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 90208.79 Da Isoelectric Point: 6.0390
>BAB52704.1 conjugal transfer protein; TrbE [Mesorhizobium japonicum MAFF 303099]
MLNLSEYRGKADRLADHLPWAALVAPGIVLNKDGSVQRTLRFRGPDLESSTEAELLGICARANNALRRLG
SGWTLFFEAERIEALGYPSSRFPDAASWLVDEERRAAFEGKVAHFESRCHLTLLSMPPPDAQARAESALV
DSHHSEGERDWRQEMARFRDETDRVLDLLSGFMPEVRPLDDAETLTYLHDTISTRRHPVAVPETPMYLDG
ILVDVPLTGGLEPMLGEQHLRTLTILGFPNLTRPGILDALNHQDFAYRWMTRFIPLDKTEATKTLTRLRR
QWFAKRKSIVAILREVVSNEPVPLVDSDADNKALDADEALQALGGDHVGFGYLTATVTVWDEDRQAAAEK
LRAVERIINGLGFTTIRESVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAEV
TGREAPPLLIAETSGSTPFRLSSHVEDVGHMLIVGPTGAGKSVLLALIALQFRRYAGAQVYVFDKGNSAR
AATLAMGGEHHVLGSDGALAFQPLRNIGDQGRRSWAAEWIAGLIAHENVTLTPEVKEAIWSALNSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHSQSAVLPVLTYLF
HRLEERFDGRPTLLMLDEAWVYLDNPLFAARIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQARTAYERFGLNDRQIELIARATPKRHYYLQSRCGNRLFELGLGPIALALCGASGPA
AQSLIDRVLSKHGQDSFVFRFLGARGLDWAAELLQQFPQPTKEQSP
MLNLSEYRGKADRLADHLPWAALVAPGIVLNKDGSVQRTLRFRGPDLESSTEAELLGICARANNALRRLG
SGWTLFFEAERIEALGYPSSRFPDAASWLVDEERRAAFEGKVAHFESRCHLTLLSMPPPDAQARAESALV
DSHHSEGERDWRQEMARFRDETDRVLDLLSGFMPEVRPLDDAETLTYLHDTISTRRHPVAVPETPMYLDG
ILVDVPLTGGLEPMLGEQHLRTLTILGFPNLTRPGILDALNHQDFAYRWMTRFIPLDKTEATKTLTRLRR
QWFAKRKSIVAILREVVSNEPVPLVDSDADNKALDADEALQALGGDHVGFGYLTATVTVWDEDRQAAAEK
LRAVERIINGLGFTTIRESVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAEV
TGREAPPLLIAETSGSTPFRLSSHVEDVGHMLIVGPTGAGKSVLLALIALQFRRYAGAQVYVFDKGNSAR
AATLAMGGEHHVLGSDGALAFQPLRNIGDQGRRSWAAEWIAGLIAHENVTLTPEVKEAIWSALNSLATAP
AQERTLTGLSVLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHSQSAVLPVLTYLF
HRLEERFDGRPTLLMLDEAWVYLDNPLFAARIREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQARTAYERFGLNDRQIELIARATPKRHYYLQSRCGNRLFELGLGPIALALCGASGPA
AQSLIDRVLSKHGQDSFVFRFLGARGLDWAAELLQQFPQPTKEQSP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 5220585..5229642
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_5014 | 5216029..5216304 | - | 276 | BAB52696 | - | - |
| Locus_5015 | 5216337..5216495 | - | 159 | BAB52697 | - | - |
| Locus_5016 | 5216933..5217364 | + | 432 | BAB52698 | - | - |
| Locus_5017 | 5217849..5220056 | + | 2208 | BAB52699 | conjugal transfer protein; TraG | - |
| Locus_5018 | 5220082..5220378 | - | 297 | BAB52700 | - | - |
| Locus_5019 | 5220397..5220588 | + | 192 | BAB52701 | - | - |
| Locus_5020 | 5220585..5221553 | + | 969 | BAB52702 | conjugal transfer protein; TraB | virB11 |
| Locus_5021 | 5221565..5221873 | + | 309 | BAB52703 | conjugal transfer protein; TrbC | virB2 |
| Locus_5022 | 5222139..5224589 | + | 2451 | BAB52704 | conjugal transfer protein; TrbE | virb4 |
| Locus_5023 | 5224586..5225311 | + | 726 | BAB52705 | conjugal transfer protein; TrbJ | virB5 |
| Locus_5024 | 5225343..5226665 | + | 1323 | BAB52706 | conjugal transfer protein; TrbL | virB6 |
| Locus_5025 | 5226666..5227391 | + | 726 | BAB52707 | conjugal transfer protein; TrbF | virB8 |
| Locus_5026 | 5227418..5228416 | + | 999 | BAB52708 | conjugal transfer protein; TrbG | virB9 |
| Locus_5027 | 5228413..5229642 | + | 1230 | BAB52709 | conjugal transfer protein; TrbI | virB10 |
| Locus_5028 | 5229655..5229924 | + | 270 | BAB52710 | - | - |
| Locus_5029 | 5230462..5231811 | - | 1350 | BAB52711 | coproporphyrinogen oxidase III | - |
| Locus_5030 | 5231969..5232724 | + | 756 | BAB52712 | transcriptional regulator | - |
| Locus_5031 | 5232728..5233267 | - | 540 | BAB52713 | - | - |
Host bacterium
| ID | 238 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEMloMAF-1 | GenBank | BA000012 |
| Element size | 7036071 bp | Coordinate of oriT [Strand] | 367747..367804 [-] |
| Host bacterium | Mesorhizobium loti MAFF303099 | Coordinate of element | 4643427..5255769 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nifA, nodF, nodE, nodZ, noeL, nolK, fixT, nifZ, nifB, fixX, fixC, fixB, fixA, nifW, nifS, mLTONO_5203, nifQ, nifH, nifD, nifK, nifE, nifN, nifX, nifU, mlr6097, nodS, nodA, nodC, nodI, nodJ, nolO, nodB, nodD, nolL, mLTONO_5487, nolX, nolW, nopB, nolT, nolU, nolV, nodM, fixN, fixO, fixQ, fixP, fixG, fixI, fixS |
| Anti-CRISPR | AcrIIC1, AcrIIA9 |