Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200143 |
| Name | oriT_ICEMcSym(1284)-alpha |
| Organism | Mesorhizobium ciceri biovar biserrulae strain WSM1284 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | CP015064 (434742..434802 [-], 61 nt) |
| oriT length | 61 nt |
| IRs (inverted repeats) | 27..33, 37..43 (CCGCCTC..GAGGCGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 61 nt
AAAAAGATGTGCAGACAAACACTTCGCCGCCTCATGGAGGCGGTGCCTTTATCTTGCCTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file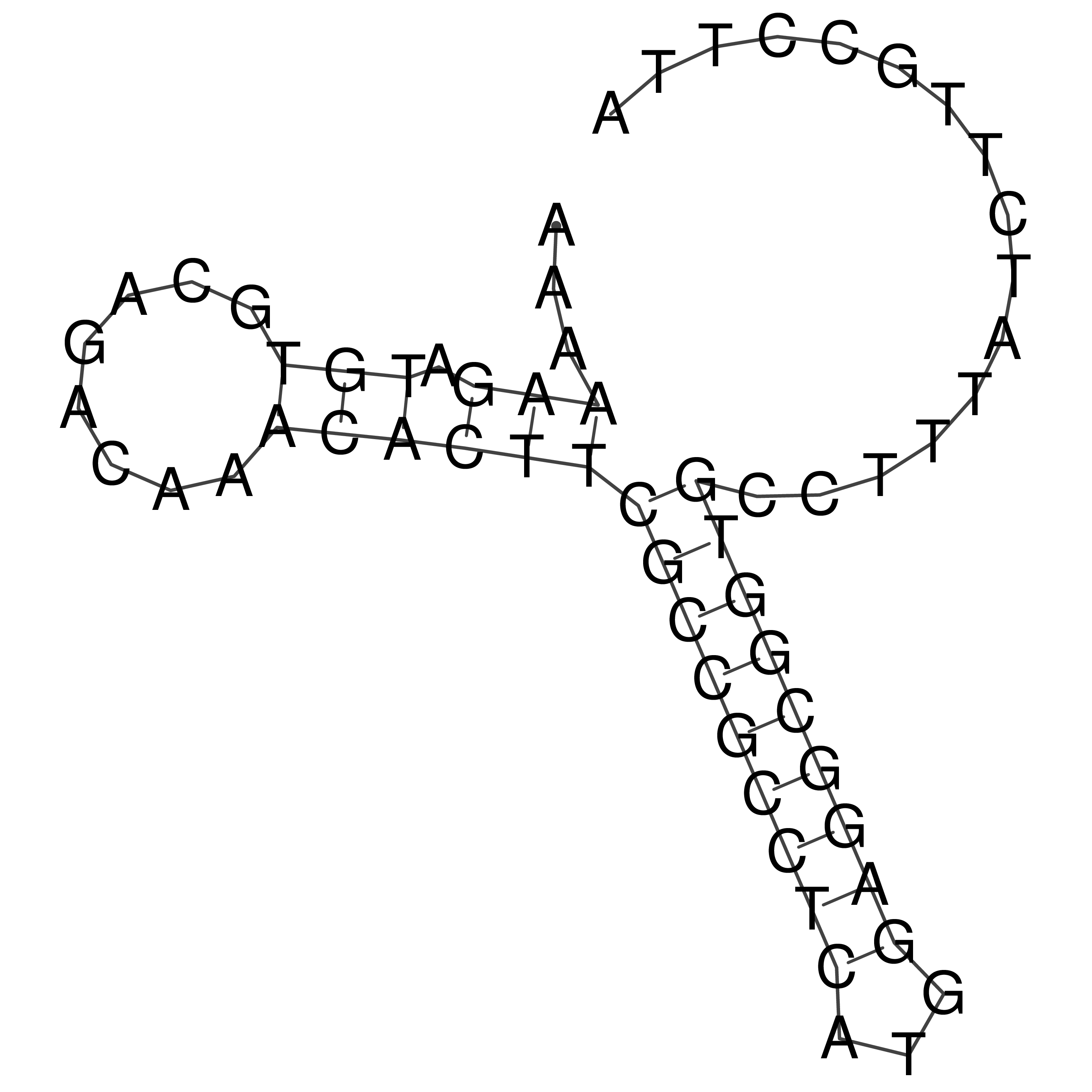
Relaxase
| ID | 14297 | GenBank | AMX98803 |
| Name | MobA_MobL_A4R29_04205_ICEMcSym(1284)-alpha |
UniProt ID | _ |
| Length | 115 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 115 a.a. Molecular weight: 13198.84 Da Isoelectric Point: 7.4484
MLPENAPEAWRDRERLWNDVEAFEVRKDAQLVREVEFALPRELSQAQGAARERDHCRNVQGRRGSLLRHM
VGSVPRCSHVQRLAAARTMGDDERRVHAMELHDALAVSCRADGHH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16584 | GenBank | AMX98970 |
| Name | t4cp2_A4R29_05350_ICEMcSym(1284)-alpha |
UniProt ID | _ |
| Length | 563 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 563 a.a. Molecular weight: 62338.73 Da Isoelectric Point: 8.6091
MPSTKTTPPNIAISIACSLALGFCAASLYATFRHGGSGEALMTFDIRAFWFETPFYLGFATPVFYRGVAI
VLATSAVVLLIQQIVLRRKLEHHGTARWARVDEMRRTGYLRRYRGVTGPVFGKTSGPFWPGYYLTNGEQP
HSLIVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKAAGDNVFKFAPLDSERRTNCYNPL
LDLIALPPQLQFTEARRLAANLITAKGEGAEGFVSGARDIFVAGILTCIERGTPTIGAVYDLFAQPGEKF
KLFAQLAMETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPAVKAATSRSDFSVYDLRRRRTC
IYLCVGPNDLEVIAPLVRLFFQQIVSILQRSLPRQDEKNEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMLIIQSLSALTGAYGEAGKQNFLSNTGLQVFMATADDETPTYISKAIGEYTFEAKSISYSQARTFDWNI
QNSHQGAPLLRPEQVRLLDEDYEIVLIKGLPPLQLRKVRYFSDRILKRIFESQTGALPEPAPLMEAHPDD
TDG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16585 | GenBank | AMX99042 |
| Name | t4cp2_A4R29_05750_ICEMcSym(1284)-alpha |
UniProt ID | _ |
| Length | 676 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 676 a.a. Molecular weight: 75085.46 Da Isoelectric Point: 6.9351
MTPTKLLIWQIAVVFAIVILGVWAATQWCAHMLGHQPSLGTPWFVVAGWPMYEPWKLFEWWYHFDAYAPE
VFDKAGALAGASGFVGCAAAIVGSLWRARQLGSVTTYGSSRWATTQEIERAGLFRPGGVFLGKLKDRYLR
HDGPEHVMAFAPTRSGKGVGLVVPTLLSWTGSAVIHDIKGENWQLTSGWRSKFSYCLLFNPTDPRSARYN
PLLEVRKGPQEVRDVQNIADILVDPEGALDRRNHWEKTSHSLLVGAVLHVLYAEEEKTLARVATFLSDPQ
RSFSATLQRMMTTNHLGTPDQPEVHPVVASAAREVLNKSENERSGVLSTAMSFLGLYRDPTVASATSACD
WRLVDLMDAERPMSLYLVVPPSDISRTKPLVRLILNQIGRRLTERLEGDPKKGRKHRLLMMLDEFPALGR
LDFFETALAFMAGYGIRAYLIAQSLNQISKAYGENSAILDNCHVRIAFSSNDDRTAKRISDALGTATELR
SMRNYAGHRLAPWLSHVMVSRQETARPLLTPGEVMQLPQNDELVLISGLPPLRAQKLRYYQDRNFTERVL
PSPVLRDGTYADCPASRPDDWTGQVRGVDSRLAAGDEIADAAIGDEGGVQQQRHPALPDNQVVLSQEADQ
GDLFKSVDDDSEPVADKRVMDRISTAARAYGINEGRGDDKDIVPGF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16586 | GenBank | AMX99046 |
| Name | t4cp2_A4R29_05775_ICEMcSym(1284)-alpha |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 89801.18 Da Isoelectric Point: 6.3596
MLNLSEYRGKADRLADHLPWAALIAPGIVLNKDGSFQRTFAFRGPDLESATEAELIGICARANNALRRLG
SGWALFFEAERIEALGYPDSRFPDAASWLVDEERRAAFQGKVAHYESRYHLTLAFMPPPDSQARAQSALV
DSHHSQGEREWRQELARFRDETGRVQDLLSGLMPEIRALDDAETLTYLHGTISTSRHPVAVPETPIYLDG
ILVDAPLTGGLEPVLGDQHLRTLTILGFPNVTRPGILDALNHQDFAYRWMTRFIPLDKTEAAKTLTRLRR
QWFAKRKSIAAILREVVTSEPVPLVDSDAENKALDADAALQALGGDHVGFGYLTTTVTVWDEDHQAAAAK
LRAVERVVNGLGFATIREGVNAVEAWLGSLPGHVYANVRQPLVHTLNLAHLMPLSSVWAGPAANEHLAKA
IQAEAPPLLFASTSGSTPFRLSTHVDDVGHMLIVGPTGAGKSVLVALFALQFRRYAGAQVYVFDKGNSAR
AAILAMGGEHHALRAHGALAFQPLRKVDDPAIRSWAAEWIAGLIAHENVTVTPEAKEAIWSALTSLATAP
AQERTLTGLSLLLQSNALKSALMPYTLDGPFGRLLDADDDRLALSDVQCFETEELMHSQGAVLPVLTNLF
HRLEARFDGRPTLLMLDEAWLYLDNPLFAARLREWLKVLRKKNVSVIFATQSLADIAGSAIAPAIIESCP
QRIFLPNDRAVEPQAREAYERFGLNERQIELVARATPKRHYYLQSRRGNRLFDLGLGVIALALCGASDPA
SQTLIDRILSEHGQDGFASHFLNARGHEWAAELLRQFPQPDMEQSS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1228364..1237794
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| A4R29_05400 | 1226075..1226338 | + | 264 | AMX98979 | hypothetical protein | - |
| A4R29_05405 | 1226395..1227084 | - | 690 | AMY03408 | DNA-binding response regulator | - |
| A4R29_05410 | 1228364..1229146 | + | 783 | AMX98980 | lytic transglycosylase | virB1 |
| A4R29_05415 | 1229161..1229526 | + | 366 | AMX98981 | type IV secretion system protein VirB2 | virB2 |
| A4R29_05420 | 1229526..1229852 | + | 327 | AMX98982 | type IV secretion system protein VirB3 | virB3 |
| A4R29_05425 | 1229852..1232221 | + | 2370 | AMX98983 | type IV secretion system protein VirB4 | virb4 |
| A4R29_05430 | 1232235..1232915 | + | 681 | AMX98984 | type IV secretion system protein VirB5 | - |
| A4R29_05435 | 1232974..1233858 | + | 885 | AMX98985 | type IV secretion system protein VirB6 | virB6 |
| A4R29_05440 | 1234040..1234753 | + | 714 | AMX98986 | type IV secretion system protein VirB8 | virB8 |
| A4R29_05445 | 1234750..1235631 | + | 882 | AMX98987 | type IV secretion system protein VirB9 | virB9 |
| A4R29_05450 | 1235628..1236770 | + | 1143 | AMX98988 | type IV secretion system protein VirB10 | virB10 |
| A4R29_05455 | 1236754..1237794 | + | 1041 | AMX98989 | type IV secretion system protein VirB11 | virB11 |
| A4R29_05460 | 1237915..1240449 | - | 2535 | AMX98990 | two-component sensor histidine kinase | - |
| A4R29_05465 | 1240628..1241380 | - | 753 | AMX98991 | carbonate dehydratase | - |
Region 2: 1300478..1309538
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| A4R29_05745 | 1295750..1297573 | + | 1824 | AMX99041 | glutamine--fructose-6-phosphate aminotransferase | - |
| A4R29_05750 | 1297920..1299950 | + | 2031 | AMX99042 | conjugal transfer protein TraG | - |
| A4R29_05755 | 1300068..1300481 | + | 414 | AMY03413 | CopG family transcriptional regulator | - |
| A4R29_05760 | 1300478..1301446 | + | 969 | AMX99043 | P-type conjugative transfer ATPase TrbB | virB11 |
| A4R29_05765 | 1301458..1301766 | + | 309 | AMX99044 | conjugal transfer protein TrbC | virB2 |
| A4R29_05770 | 1301766..1302035 | + | 270 | AMX99045 | conjugal transfer protein | virB3 |
| A4R29_05775 | 1302029..1304479 | + | 2451 | AMX99046 | conjugal transfer protein TrbE | virb4 |
| A4R29_05780 | 1304476..1305201 | + | 726 | AMX99047 | conjugal transfer protein TrbJ | virB5 |
| A4R29_05785 | 1305232..1306554 | + | 1323 | AMX99048 | conjugal transfer protein TrbL | virB6 |
| A4R29_05790 | 1306555..1307286 | + | 732 | AMX99049 | conjugal transfer protein TrbF | virB8 |
| A4R29_05795 | 1307276..1308304 | + | 1029 | AMX99050 | P-type conjugative transfer protein TrbG | virB9 |
| A4R29_05800 | 1308318..1309538 | + | 1221 | AMX99051 | conjugal transfer protein TrbI | virB10 |
| A4R29_05805 | 1309541..1309813 | + | 273 | AMX99052 | transposase | - |
| A4R29_05810 | 1310134..1311483 | - | 1350 | AMX99053 | coproporphyrinogen III oxidase | - |
| A4R29_05815 | 1311639..1312388 | + | 750 | AMX99054 | transcriptional regulator | - |
| A4R29_05820 | 1312894..1314522 | + | 1629 | AMX99055 | peptidase S41 | - |
Host bacterium
| ID | 145 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEMcSym(1284)-alpha | GenBank | CP015064 |
| Element size | 6326813 bp | Coordinate of oriT [Strand] | 434742..434802 [-] |
| Host bacterium | Mesorhizobium ciceri biovar biserrulae strain WSM1284 | Coordinate of element | 858217..1335305 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodF, nodE, nodA, nodZ, noeL, nolK, nodC, nodI, nodJ, a4R29_04430, nodB, nodD, nolL, nodU, nifA, fixU, nifZ, nifB, fixX, fixC, fixB, fixA, nifW, nifS, nifQ, nifH, nifD, nifK, nifE, nifN, nifX, nodM, fixN, fixO, fixQ, fixP, fixG, fixH, fixI, fixS |
| Anti-CRISPR | - |