Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200131 |
| Name | oriT_ICEVchChn2255 |
| Organism | Vibrio cholerae strain ICDC-2255 sequence |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | KT151660 (86649..86947 [-], 299 nt) |
| oriT length | 299 nt |
| IRs (inverted repeats) | 234..242, 254..262 (AAAGCCAAA..TTTGGCTTT) 45..50, 52..57 (CAAACG..CGTTTG) 7..12, 26..31 (TTTGGC..GCCAAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 299 nt
>oriT_ICEVchChn2255
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
GCTCTGTTTGGCGGCTGGTGACCTAGCCAAAAAAATTGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTTGCCAAACGGTTTGTATCTTCATGGCGATACGTCTTTTTAGGTGTTTTTAAGTGAAATCAGCCTGTATCCCTTGTCGGGTATGGGATTGAGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file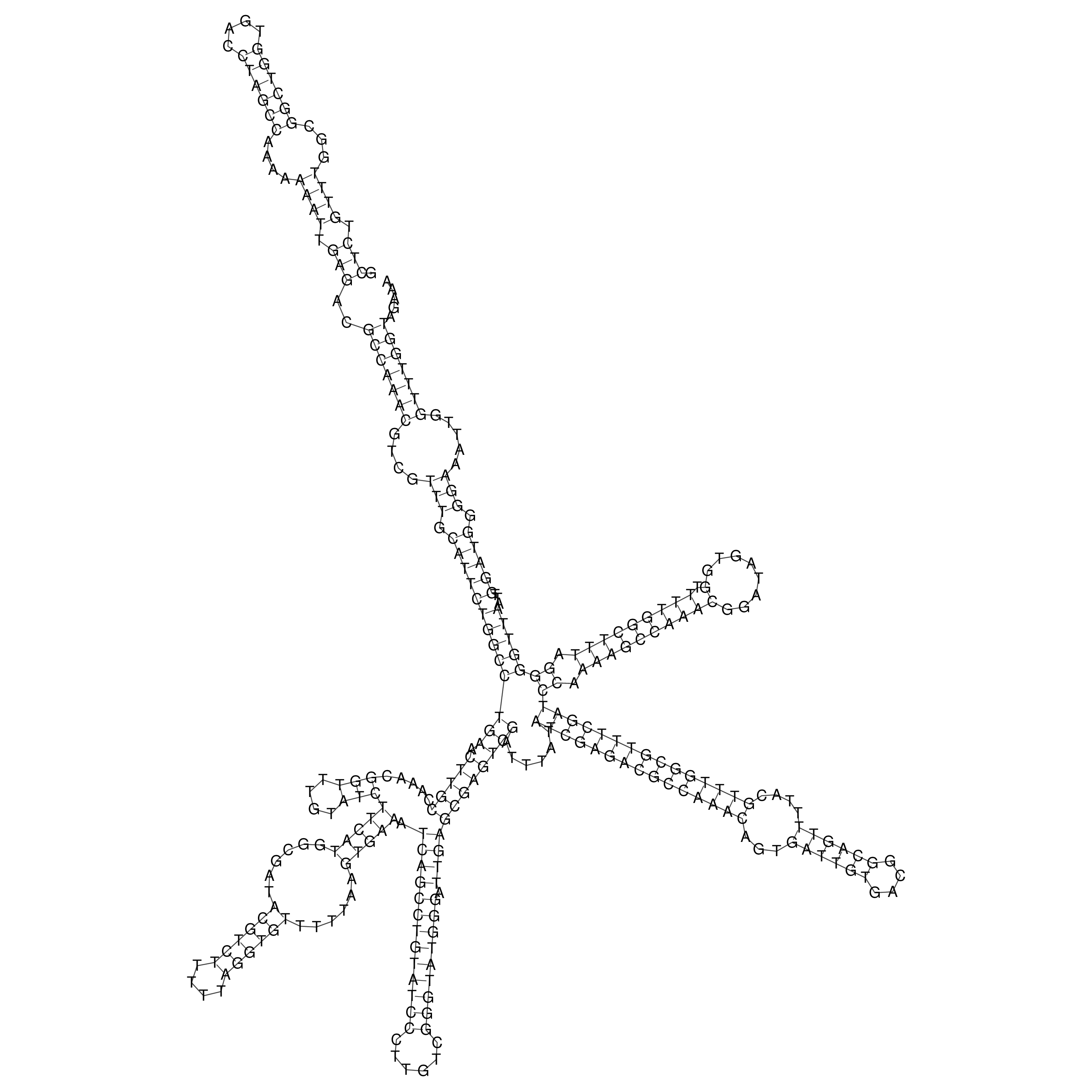
Relaxase
| ID | 14284 | GenBank | ALP44909 |
| Name | traI_-_ICEVchChn2255 |
UniProt ID | _ |
| Length | 716 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 716 a.a. Molecular weight: 79789.91 Da Isoelectric Point: 5.2251
>ALP44909.1 conjugative transfer protein relaxase [Vibrio cholerae]
MFKNLFFQTKALPELSSQLDTEIPRYPPFLKGLPAASPEDLQSTQDELITKLRQVLGFNLRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNTIGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTNLYLVWKSAAKEIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDGKPIWLPMLHISEADLLFSSNVPSGVRLFSKSEWEATQQTQAEPQSRSSEQPGL
SEASSSIELNNSAESPSTKSSEQDDERRLASDVNHLQATENAPGDECEKPNNSYDGAISNNVNQHDAEAL
NLPESLAWLPEASSALVMVDEQILVRYPDAVRHWCAPRKLLAELSRLDWLEIDPANPTRKARTVTTKGGV
QEQGLLLKVSISKGLTALIDLPKQDAESAAAIQNEEASPRPSRTETTNARAKEPSKRAECKQKPIAPNAN
SSIDPKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAER
MFKNLFFQTKALPELSSQLDTEIPRYPPFLKGLPAASPEDLQSTQDELITKLRQVLGFNLRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNTIGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTNLYLVWKSAAKEIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDGKPIWLPMLHISEADLLFSSNVPSGVRLFSKSEWEATQQTQAEPQSRSSEQPGL
SEASSSIELNNSAESPSTKSSEQDDERRLASDVNHLQATENAPGDECEKPNNSYDGAISNNVNQHDAEAL
NLPESLAWLPEASSALVMVDEQILVRYPDAVRHWCAPRKLLAELSRLDWLEIDPANPTRKARTVTTKGGV
QEQGLLLKVSISKGLTALIDLPKQDAESAAAIQNEEASPRPSRTETTNARAKEPSKRAECKQKPIAPNAN
SSIDPKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAER
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16554 | GenBank | ALP44891 |
| Name | traC_-_ICEVchChn2255 |
UniProt ID | _ |
| Length | 799 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 799 a.a. Molecular weight: 90098.71 Da Isoelectric Point: 6.2880
>ALP44891.1 plasmid conjugative transfer pilus assembly protein trac [Vibrio cholerae]
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAAISAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTGEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRNSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAAISAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTGEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRNSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16555 | GenBank | ALP44908 |
| Name | traD_-_ICEVchChn2255 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67134.99 Da Isoelectric Point: 6.4440
>ALP44908.1 plasmid conjugative transfer protein [Vibrio cholerae]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQMG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPERSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSGMALYRLPEAYRLYKLQMG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKQTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPERSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 29105..53242
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_29 | 24903..25046 | - | 144 | ALP44879 | hypothetical protein | - |
| Locus_30 | 25128..25946 | - | 819 | ALP44880 | recombination protein BET | - |
| Locus_31 | 26026..26445 | - | 420 | ALP44881 | single-stranded DNA-binding protein | - |
| Locus_32 | 26461..26787 | - | 327 | ALP44882 | hypothetical protein | - |
| Locus_33 | 27155..27757 | + | 603 | ALP44883 | hypothetical protein | - |
| Locus_34 | 27848..28510 | - | 663 | ALP44884 | hypothetical protein | - |
| Locus_35 | 28541..28873 | - | 333 | ALP44885 | hypothetical protein | - |
| Locus_36 | 29105..32797 | - | 3693 | ALP44886 | plasmid conjugative transfer protein | traN |
| Locus_37 | 32800..33828 | - | 1029 | ALP44887 | plasmid conjugative transfer pilus assembly protein trau | traU |
| Locus_38 | 33812..34936 | - | 1125 | ALP44888 | plasmid conjugative transfer pilus assembly protein traw | traW |
| Locus_39 | 34947..35459 | - | 513 | ALP44889 | conjugative signal peptidase | - |
| Locus_40 | 35443..35790 | - | 348 | ALP44890 | hypothetical protein | - |
| Locus_41 | 35783..38182 | - | 2400 | ALP44891 | plasmid conjugative transfer pilus assembly protein trac | virb4 |
| Locus_42 | 38182..38874 | - | 693 | ALP44892 | thiol:disulfide protein | trbB |
| Locus_43 | 39253..39942 | - | 690 | ALP44893 | ync | - |
| Locus_44 | 39935..40768 | - | 834 | ALP44894 | ynd | - |
| Locus_45 | 40946..41332 | - | 387 | ALP44895 | conjugative transfer protein | - |
| Locus_46 | 41329..41979 | - | 651 | ALP44896 | conjugative transfer protein | traV |
| Locus_47 | 41976..43265 | - | 1290 | ALP44897 | plasmid conjugative transfer pilus assembly protein trab | traB |
| Locus_48 | 43268..44164 | - | 897 | ALP44898 | plasmid conjugative transfer pilus assembly protein | traK |
| Locus_49 | 44148..44774 | - | 627 | ALP44899 | plasmid conjugative transfer pilus assembly protein trae | traE |
| Locus_50 | 44771..45052 | - | 282 | ALP44900 | plasmid conjugative transfer pilus assembly protein tral | traL |
| Locus_51 | 45341..45913 | + | 573 | ALP44901 | hypothetical protein | - |
| Locus_52 | 45992..46687 | - | 696 | ALP44902 | ISPsy4 transposition helper protein | virB11 |
| Locus_53 | 46763..48034 | - | 1272 | ALP44903 | hypothetical protein | - |
| Locus_54 | 47994..48242 | - | 249 | ALP44904 | hypothetical protein | - |
| Locus_55 | 48402..49958 | - | 1557 | ALP44905 | transposase | - |
| Locus_56 | 50230..50865 | - | 636 | ALP44906 | conjugative transfer protein | tfc7 |
| Locus_57 | 50852..51412 | - | 561 | ALP44907 | conjugative transfer protein | - |
| Locus_58 | 51422..53242 | - | 1821 | ALP44908 | plasmid conjugative transfer protein | virb4 |
| Locus_59 | 53291..55441 | - | 2151 | ALP44909 | conjugative transfer protein relaxase | - |
| Locus_60 | 55561..56034 | - | 474 | ALP44910 | hypothetical protein | - |
| Locus_61 | 56034..56315 | - | 282 | ALP44911 | hypothetical protein | - |
| Locus_62 | 56409..57263 | - | 855 | ALP44912 | mrr restriction system protein | - |
Host bacterium
| ID | 133 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEVchChn2255 | GenBank | KT151660 |
| Element size | 92592 bp | Coordinate of oriT [Strand] | 86649..86947 [-] |
| Host bacterium | Vibrio cholerae strain ICDC-2255 sequence | Coordinate of element | 1..92592 |
Cargo genes
| Drug resistance gene | dfrA1, sul2, aph(3'')-Ib, aph(6)-Id, tet(59) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |