Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200124 |
| Name | oriT_ICEValHN437 |
| Organism | Vibrio alginolyticus strain HN437 sequence |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | KT072771 (3488..3786 [+], 299 nt) |
| oriT length | 299 nt |
| IRs (inverted repeats) | 45..50, 52..57 (CAAACG..CGTTTG) 7..12, 26..31 (TTTGGC..GCCAAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 299 nt
>oriT_ICEValHN437
GCTCTGTTTGGCGGCGGATGAATGAGCCAAAAAAATCGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTCGCCAAACGGTTTGTATCTTCATGACGATACGTCTTTTTAGGCGTTTTTAAGTGAAATCGGGCTGTATCCCTTGTCAGATATGGGATTGCGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGCTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
GCTCTGTTTGGCGGCGGATGAATGAGCCAAAAAAATCGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTCGCCAAACGGTTTGTATCTTCATGACGATACGTCTTTTTAGGCGTTTTTAAGTGAAATCGGGCTGTATCCCTTGTCAGATATGGGATTGCGCGAGTCGATTTATATCGAGACGCCAAACAGTGATTGCTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file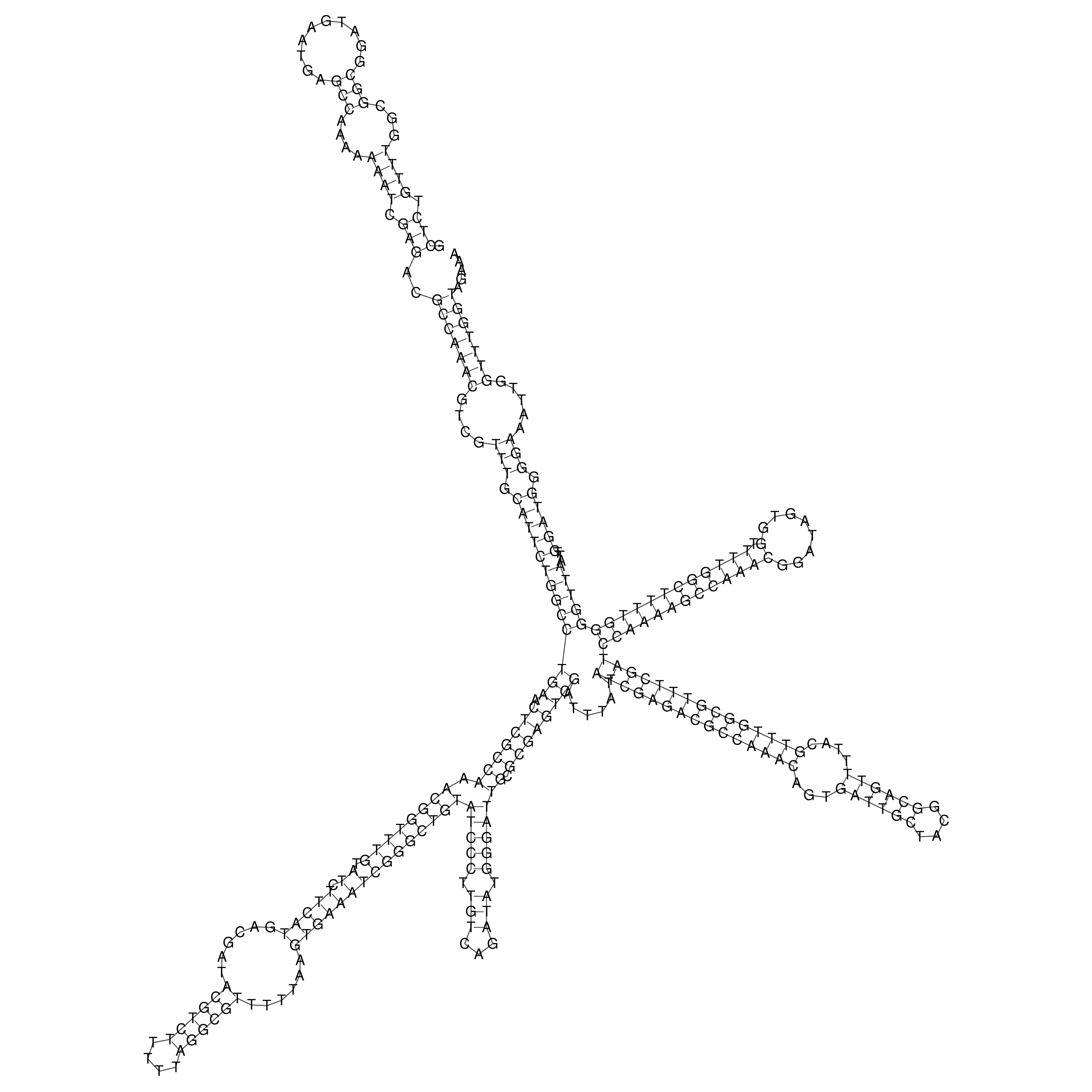
Relaxase
| ID | 14277 | GenBank | ALF35094 |
| Name | traI_ICEValHN437_020_ICEValHN437 |
UniProt ID | _ |
| Length | 716 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 716 a.a. Molecular weight: 79655.68 Da Isoelectric Point: 5.0979
>ALF35094.1 Conjugative transfer protein, TraI [Vibrio alginolyticus]
MFKNLFFQTKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVIGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDTPIWLPMLHIVEADLLFSSNVPSSVALFSKSEWEATQQTQAEPQCRSSEHSDL
PEASSSIELSNSAESPSSKSSDQDDELRLASDVNHLQATENAPGDECEKPNNSYDGAISNNVNQQDAEAL
NLPESLAWLPEASSALIMVGEQLLIRYPDAVRPWCAPRKLLAELSRLDWLELDPANPTRKARTITTKDGV
QEQGLLLKVSISKGLTALIDISKHDTESAAAIQNKEASQRPSRTETTNAQAKESATRAERKQKPIAANAN
SSTDPKHAQRQQMVSFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDEC
QFDEGETVLFTAHAKR
MFKNLFFQTKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVIGFNQRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWIRKSNQSTHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLATKSA
SNERYESLAPEVLIKDDTPIWLPMLHIVEADLLFSSNVPSSVALFSKSEWEATQQTQAEPQCRSSEHSDL
PEASSSIELSNSAESPSSKSSDQDDELRLASDVNHLQATENAPGDECEKPNNSYDGAISNNVNQQDAEAL
NLPESLAWLPEASSALIMVGEQLLIRYPDAVRPWCAPRKLLAELSRLDWLELDPANPTRKARTITTKDGV
QEQGLLLKVSISKGLTALIDISKHDTESAAAIQNKEASQRPSRTETTNAQAKESATRAERKQKPIAANAN
SSTDPKHAQRQQMVSFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDEC
QFDEGETVLFTAHAKR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16540 | GenBank | ALF35152 |
| Name | traD_ICEValHN437_021_ICEValHN437 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67068.90 Da Isoelectric Point: 6.6974
>ALF35152.1 Conjugative transfer protein, TraD [Vibrio alginolyticus]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEILTELPPEPFWWMTGISSGMALYRLPEAYRLHKLQKG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKKTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEILTELPPEPFWWMTGISSGMALYRLPEAYRLHKLQKG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKKTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAAGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16541 | GenBank | ALF35093 |
| Name | traC_ICEValHN437_037_ICEValHN437 |
UniProt ID | _ |
| Length | 800 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 800 a.a. Molecular weight: 90283.82 Da Isoelectric Point: 5.8195
>ALF35093.1 Conjugative transfer pilus assembly protein, TraC [Vibrio alginolyticus]
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLLGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAESI
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMNLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHDDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGFDE
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLLGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWLAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAESI
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMNLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHDDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGFDE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 22133..42213
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ICEValHN437_019 | 18985..19839 | + | 855 | ALF35181 | Mrr restriction system protein | - |
| ICEValHN437_020 | 19934..22084 | + | 2151 | ALF35094 | Conjugative transfer protein, TraI | - |
| ICEValHN437_021 | 22133..23953 | + | 1821 | ALF35152 | Conjugative transfer protein, TraD | virb4 |
| ICEValHN437_022 | 23963..24523 | + | 561 | ALF35170 | Conjugative transfer protein, S091 | - |
| ICEValHN437_023 | 24510..25145 | + | 636 | ALF35088 | Conjugative transfer protein, traJ | tfc7 |
| ICEValHN437_024 | 25119..25238 | - | 120 | ALF35137 | Hypothetical protein | - |
| ICEValHN437_025 | 25283..26389 | + | 1107 | ALF35110 | Fic family protein | - |
| ICEValHN437_026 | 26445..26579 | - | 135 | ALF35175 | Hypothetical protein | - |
| ICEValHN437_027 | 26591..26881 | - | 291 | ALF35171 | HigA protein | - |
| ICEValHN437_028 | 27114..27395 | + | 282 | ALF35174 | Conjugative transfer pilus assembly protein, TraL | traL |
| ICEValHN437_029 | 27392..28018 | + | 627 | ALF35091 | Conjugative transfer pilus assembly protein, TraE | traE |
| ICEValHN437_030 | 28002..28898 | + | 897 | ALF35101 | Conjugative transfer pilus assembly protein, TraK | traK |
| ICEValHN437_031 | 28901..30190 | + | 1290 | ALF35120 | Conjugative transfer pilus assembly protein, TraB | traB |
| ICEValHN437_032 | 30187..30837 | + | 651 | ALF35147 | Conjugative transfer protein, TraV | traV |
| ICEValHN437_033 | 30834..31220 | + | 387 | ALF35115 | Conjugative transfer protein, TraA | - |
| ICEValHN437_034 | 31261..31767 | - | 507 | ALF35113 | Acetyltransferase | - |
| ICEValHN437_035 | 31758..32024 | - | 267 | ALF35143 | Hypothetical protein | - |
| ICEValHN437_036 | 32440..33132 | + | 693 | ALF35178 | Thiol:disulfide involved in conjugative transfer, DsbC | trbB |
| ICEValHN437_037 | 33133..35535 | + | 2403 | ALF35093 | Conjugative transfer pilus assembly protein, TraC | virb4 |
| ICEValHN437_038 | 35528..35875 | + | 348 | ALF35159 | Conjugative transfer protein 345 | - |
| ICEValHN437_039 | 35859..36371 | + | 513 | ALF35119 | Conjugative signal peptidase, TrhF | - |
| ICEValHN437_040 | 36382..37506 | + | 1125 | ALF35158 | Conjugative transfer pilus assembly protein, TraW | traW |
| ICEValHN437_041 | 37766..38518 | + | 753 | ALF35141 | Conjugative transfer pilus assembly protein, TraU | traU |
| ICEValHN437_042 | 38521..42213 | + | 3693 | ALF35155 | Conjugative transfer protein, TraN | traN |
| ICEValHN437_043 | 42445..42777 | + | 333 | ALF35162 | Hypothetical protein | - |
| ICEValHN437_044 | 42808..43470 | + | 663 | ALF35106 | Hypothetical protein | - |
| ICEValHN437_045 | 43561..44163 | - | 603 | ALF35123 | Hypothetical protein, S063 | - |
| ICEValHN437_046 | 44531..44857 | + | 327 | ALF35134 | Hypothetical protein, S089 | - |
| ICEValHN437_047 | 44873..45292 | + | 420 | ALF35105 | Single-stranded DNA-binding protein, Ssb | - |
| ICEValHN437_048 | 45371..46189 | + | 819 | ALF35161 | Recombination protein, Bet | - |
| ICEValHN437_049 | 46272..46415 | + | 144 | ALF35098 | Hypothetical protein, OrfZ | - |
Host bacterium
| ID | 126 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEValHN437 | GenBank | KT072771 |
| Element size | 94290 bp | Coordinate of oriT [Strand] | 3488..3786 [+] |
| Host bacterium | Vibrio alginolyticus strain HN437 sequence | Coordinate of element | 1..94290 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | ugd |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |