Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200104 |
| Name | oriT_ICEPmiCHN1809 |
| Organism | Proteus mirabilis strain TJ1809 integrative and |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | KX243413 (3888..4186 [+], 299 nt) |
| oriT length | 299 nt |
| IRs (inverted repeats) | 45..50, 52..57 (CAAACG..CGTTTG) 7..12, 26..31 (TTTGGC..GCCAAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 299 nt
>oriT_ICEPmiCHN1809
GCTCTGTTTGGCGGCGGATGACCTAGCCAAAAAATTCGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTCGCCAAACGGTTTGTATCTTCATGACGATACGTCTTTTTAGGCGTTTTTAAGTGAAATCGGGCTGTATCCCTTGTCAGGTATGGGATTGCGCGAGTTGATTTATATCGAGACGCCAAACAGTGATTGTTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
GCTCTGTTTGGCGGCGGATGACCTAGCCAAAAAATTCGAGACGCCAAACGTCGTTTGCATTCTGGCCTGAACTCGCCAAACGGTTTGTATCTTCATGACGATACGTCTTTTTAGGCGTTTTTAAGTGAAATCGGGCTGTATCCCTTGTCAGGTATGGGATTGCGCGAGTTGATTTATATCGAGACGCCAAACAGTGATTGTTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGATAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAAATTGGTTTGGTAGAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file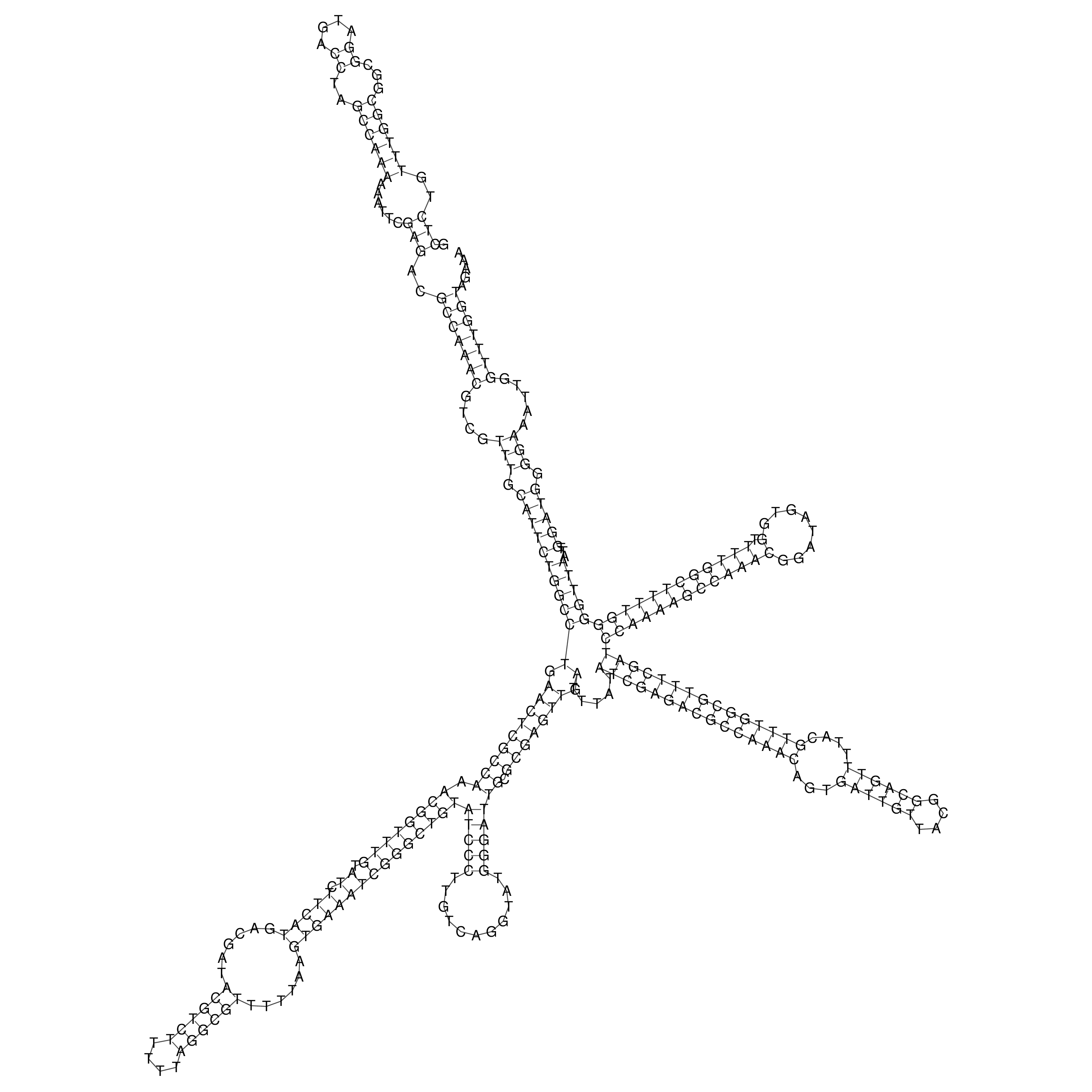
Relaxase
| ID | 14257 | GenBank | APO17569 |
| Name | traI_-_ICEPmiCHN1809 |
UniProt ID | _ |
| Length | 716 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 716 a.a. Molecular weight: 79478.67 Da Isoelectric Point: 5.2745
>APO17569.1 Conjugative transfer protein TraI, relaxase [Proteus mirabilis]
MFKNLFFQTKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNLRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWVQKSNQSIHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLAIKSA
SNERYESFAPEVLIKDGKPIWLPMLHISEADLLFSSNLPSGVTLFNKSQWEATQQTQAEPLSRSSEHSDL
PEASTSIELSNSAESPSSKSSEQDDGLSRASDVNNPQATENAPGDGCEKPNNSYDGAISNNVNQHDAEAL
NLPESLAWLPEASSALVVVGEQILIRYPDAVRPWCAPRKLLAELSQLDWLELDPANPTRKARSITTNDGV
QEQGLLLKVSISKVLTALIDLPKQDAESAAAIQNEEASQRPSRTKITNAQAKEPAKRAERKQKPIAPNAN
SSTDLKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAKR
MFKNLFFQTKALPELSSQLDAEIPRYPPFLKGLPAASPEDLQSTQDELIAKLRQVLGFNLRDFQRLIQPC
IDHLAAYVHLLPASEHHHHSGAGGLLRHSLEVAFWAAQAAEGIIFVASGTPVEKKELEPRWRVAAALGGL
FHDIGKPVSDLSITDEDGRYQWNPFLETLSQWTTNNSIERYFIRWRDGRCKRHEQFSILVLNRVMTPELL
AWLTQPGPEILQAMLEAIGNTDPEHVLSKLVIEADQTSVQRDLKAQRISVDDNALGVPVERYLLDAMRRL
LASSQWLVNQRDARVWVQKSNQSIHLYLVWKSAAKDIIELLAKDKIPGIPRDPDTLADILIERGLAIKSA
SNERYESFAPEVLIKDGKPIWLPMLHISEADLLFSSNLPSGVTLFNKSQWEATQQTQAEPLSRSSEHSDL
PEASTSIELSNSAESPSSKSSEQDDGLSRASDVNNPQATENAPGDGCEKPNNSYDGAISNNVNQHDAEAL
NLPESLAWLPEASSALVVVGEQILIRYPDAVRPWCAPRKLLAELSQLDWLELDPANPTRKARSITTNDGV
QEQGLLLKVSISKVLTALIDLPKQDAESAAAIQNEEASQRPSRTKITNAQAKEPAKRAERKQKPIAPNAN
SSTDLKHAQRQQMVNFVKDLPILLTDGDYPDVDHSADGIRVTIQTLRQVANEHGIPAGQLLRGISASDQC
QFDEGETVLFTAHAKR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16500 | GenBank | APO17570 |
| Name | traD_-_ICEPmiCHN1809 |
UniProt ID | _ |
| Length | 606 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 606 a.a. Molecular weight: 67129.01 Da Isoelectric Point: 6.6974
>APO17570.1 IncF plasmid conjugative transfer protein TraD [Proteus mirabilis]
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSAMALYRLPEAYRLHKLQKG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKKTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAVGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
MKENAYEMPWRTNYEAMAAAGWLVGATGAIAAEMLTELPPEPFWWMTGISSAMALYRLPEAYRLHKLQKG
LKGKPLAFMELSHLQKVMAKHPDELWLGYGFEWDQRHAQRAYEILKRDKKTLLNQGHGKQMGSTWIHGVE
PKEEDVYQPVGHTEGHTLIVGTTGAGKTRCFDAMITQAILRNEAVIIIDPKGDKELKDNAQRACIAVGSP
ERFVYFHPGFPEHSVRLNPLRNFNRGTEIASRIAALIPSETGADPFKAFGQMALNNIVQGLLLTSQRPDL
KTLRRFLEGGPEGLVVKAVTAWGEQVYPNFSVEIKRFTEKANTLAKQAMAMLLFYYERIQPVAANTDLEG
LLSMFEHDRTHYSKMVASLMPVLNMLTSSELGPLLSPIANDVDDSRLITDSGRIINNAQVAYIGLDSLTD
AMVGSAIGSLLLSDLTAVAGDRYNYGVENRPVNIFIDEAAEVVNDPFIQLLNKGRGAKMRCVIATQTFAD
FAARTGSEAKARQVLGNINNLIALRVMDAETQQYITDNLPKTRLQYIMQTQGMSSNSDSPALFTGNHGER
LMEEEGDMFPPQLLGQLPNLEYIAKLSGGRVIKGRIPILTSSTQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16501 | GenBank | APO17582 |
| Name | traC_-_ICEPmiCHN1809 |
UniProt ID | _ |
| Length | 799 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 799 a.a. Molecular weight: 90171.75 Da Isoelectric Point: 5.9735
>APO17582.1 IncF plasmid conjugative transfer pilus assembly protein TraC [Proteus mirabilis]
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWIAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
MKASLYSGQRASELLPVLAYSDDEQLFFMEDQSVGFGFLCDPLPGGDESVADRVNVLLNNDWPKDTLLQF
GLYASPDIQTDLQRMMGLRHRQSDPLLRASIRKRADFLDGGTVQPIEESTQTQVRNFQLIVTCKLPLESP
IPTERELSRASALRASFSQALATVGFRVTEMTDRNWIAALSAQLNWGKDASWRNPSPIRSEADKPLREQV
LDYDRAIKVDSQGLMLGDYRVKTLSFKRLPERIWFGHAASFAGDMMTGSRGLRGSFLLNVTIHFPSAEAM
RSRLETKRQWAVNQAYGPMLKFVPVLAAKKKGFDVLFEALQEGDRPIRANMTLTLFSPTEEASISSVSNA
RTYFKELGFELMEDKYFCLPIFLNALPFGADRQAMNDLFRFKTMATRHVIPLLPLFADWKGTGTPVINFV
SRNGQIMSVSLYDSGSNYNCCIAAQSGSGKSFLVNEIISSYLSEGGQCWVIDVGRSYEKLCEVYDGEFLQ
FGRDSGICLNPFEIVEDYDEEADVLVGLLAAMAAPTQSLTDFQMANLKRQTRELWEKKGRAMLVDDVAEA
LKNHEDRRVQDVGEQLYPFTTQGEYGRFFNGHNNIRFKNRFTVLELEELKGRKHLQQVVLLQLIYQIQQE
MYLGERDRRKIVFIDEAWDLLTQGDVGKFIETGYRRFRKYGGSAVTVTQSVNDLYDSPTGKAIAENSANM
YLLGQKAETINALKKEGRLPLGEGGYEYLKTVHTVTGVYSEIFFITEMGTGIGRLIVDPFHKLLYSSRAE
DVNAIKQLTRKGLSVADAISQLLKERGYE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 23294..42445
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_17 | 20131..21000 | + | 870 | APO17568 | Mrr restriction endonuclease | - |
| Locus_18 | 21095..23245 | + | 2151 | APO17569 | Conjugative transfer protein TraI, relaxase | - |
| Locus_19 | 23294..25114 | + | 1821 | APO17570 | IncF plasmid conjugative transfer protein TraD | virb4 |
| Locus_20 | 25124..25684 | + | 561 | APO17571 | Conjugative transfer protein 234 | - |
| Locus_21 | 25671..26306 | + | 636 | APO17572 | Conjugative transfer protein s043 | tfc7 |
| Locus_22 | 26333..26920 | - | 588 | APO17573 | hypothetical protein | - |
| Locus_23 | 27209..27490 | + | 282 | APO17574 | IncF plasmid conjugative transfer pilus assembly protein TraL | traL |
| Locus_24 | 27487..28113 | + | 627 | APO17575 | IncF plasmid conjugative transfer pilus assembly protein TraE | traE |
| Locus_25 | 28097..28993 | + | 897 | APO17576 | IncF plasmid conjugative transfer pilus assembly protein TraK | traK |
| Locus_26 | 28996..30285 | + | 1290 | APO17577 | IncF plasmid conjugative transfer pilus assembly protein TraB | traB |
| Locus_27 | 30360..30932 | + | 573 | APO17578 | Conjugative transfer protein TraV | traV |
| Locus_28 | 30929..31315 | + | 387 | APO17579 | Conjugative transfer protein TraA | - |
| Locus_29 | 31396..32334 | - | 939 | APO17580 | TIORF34 protein | - |
| Locus_30 | 32676..33368 | + | 693 | APO17581 | Thiol:disulfide involved in conjugative transfer | trbB |
| Locus_31 | 33368..35767 | + | 2400 | APO17582 | IncF plasmid conjugative transfer pilus assembly protein TraC | virb4 |
| Locus_32 | 35760..36107 | + | 348 | APO17583 | Conjugative transfer protein 345 | - |
| Locus_33 | 36091..36603 | + | 513 | APO17584 | Conjugative signal peptidase TrhF | - |
| Locus_34 | 36614..37738 | + | 1125 | APO17585 | IncF plasmid conjugative transfer pilus assembly protein TraW | traW |
| Locus_35 | 37770..38750 | + | 981 | APO17586 | IncF plasmid conjugative transfer pilus assembly protein TraU | traU |
| Locus_36 | 38753..42445 | + | 3693 | APO17587 | IncF plasmid conjugative transfer protein TraN | traN |
| Locus_37 | 42755..43645 | + | 891 | APO17588 | putative integrase | - |
| Locus_38 | 43642..44694 | + | 1053 | APO17589 | transposase | - |
Host bacterium
| ID | 106 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEPmiCHN1809 | GenBank | KX243413 |
| Element size | 76218 bp | Coordinate of oriT [Strand] | 3888..4186 [+] |
| Host bacterium | Proteus mirabilis strain TJ1809 integrative and | Coordinate of element | 1..76218 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |