Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200095 |
| Name | oriT_CTn341 |
| Organism | Bacteroides fragilis |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | AY515263 (19315..19514 [+], 200 nt) |
| oriT length | 200 nt |
| IRs (inverted repeats) | 40..56, 61..77 (CTTTTCAGCCGCTCAAA..TTTGAGCAGCCACGAAA) |
| Location of nic site | 79..80 |
| Conserved sequence flanking the nic site |
AAAAG|CACTTGC |
| Note |
oriT sequence
Download Length: 200 nt
CCAACGGATTTTTATGTTCACCTGAACATAGCAAGATGTCTTTTCAGCCGCTCAAAATATTTTGAGCAGCCACGAAAAGCACTTGCCCTTGCAGGGGGCGGGCTTTCCCTCCGAAGTCGGGAAGCCTTTCAGAACTGCGGTGCTGTAATCCGAAGCGGCGGCTTATGCCGTCAATCCGCTAAATCCAAAGTAATATGAAA
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file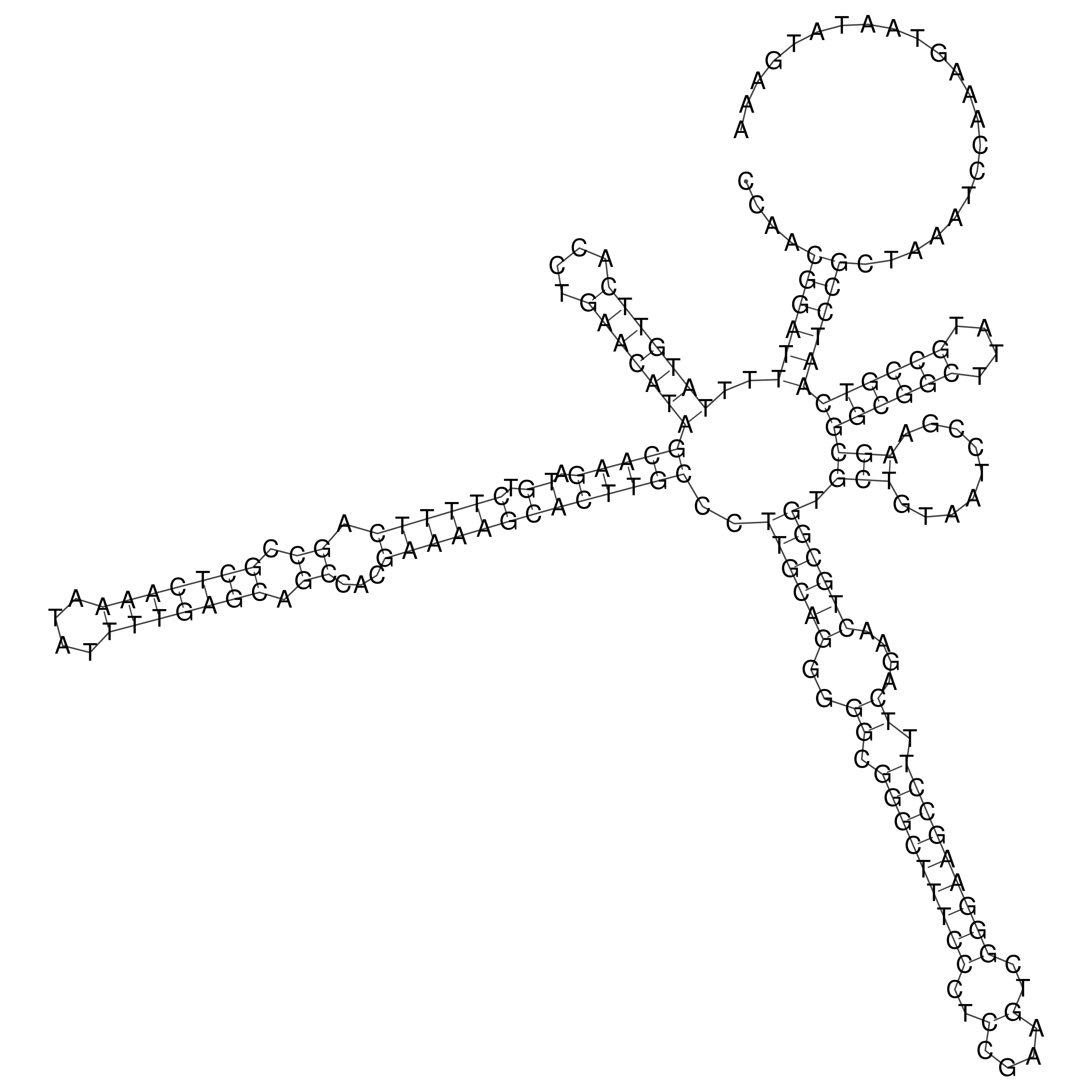
Reference
[1] Lindsay Peed et al. (2010) Genetic and functional analyses of the mob operon on conjugative transposon CTn341 from Bacteroides spp. Journal of bacteriology. 192(18):4643-50. [PMID:20639338]
Relaxase
| ID | 1099 | GenBank | AAS83499 |
| Name | mob341_CTn341 |
UniProt ID | Q56VF2 |
| Length | 415 a.a. | PDB ID | |
| Note | |||
Relaxase protein sequence
Download Length: 415 a.a. Molecular weight: 46563.08 Da Isoelectric Point: 9.8931
MVAKISVGSSLYGAIAYNGEKINEAQGRLLTTNRIYNDGSGTVDIGKAMEGFLTFLPPQMKIEKPVVHIS
LNPHPEDVLTDIELQNIAREYLEKLGFGNQPYLVFKHEDIDRHHLHIVTVNVDENGKRLNRDFLYRRSDR
IRRELEQKYGLHPAERKNQRLDNPLRKVAASAGDVKKQVGNTVKALNGQYRFQTMGEYRALLSLYNMTVE
EARGNVRGREYHGLVYSVTDDKGNKVGNPFKSSLFGKSAGYEAVQKKFVRSKSEIKDRKLADMTKRTVLS
VLQGTYDKDKFVSQLKEKGIDTVLRYTEEGRIYGATFIDHRTGCVLNGSRMGKELSANALQEHFTLPYAG
QPPIPLSIPVDAADKAHGQTAYDSEDISGGMGLLTPEGPAVDAEEEAFIRAMKRKKKKKRKGLGM
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q56VF2 |
Reference
[1] Lindsay Peed et al. (2010) Genetic and functional analyses of the mob operon on conjugative transposon CTn341 from Bacteroides spp. Journal of bacteriology. 192(18):4643-50. [PMID:20639338]
Auxiliary protein
| ID | 394 | GenBank | AAS83500 |
| Name | mobC341_CTn341 |
UniProt ID | Q56VF1 |
| Length | 672 a.a. | PDB ID | _ |
| Note | |||
Auxiliary protein sequence
Download Length: 672 a.a. Molecular weight: 77086.10 Da Isoelectric Point: 7.1001
MSQQEDDLRALAKIMDFLRAVSIILVVMNVYWFCYEAIRLWGVNIGVVDKILLNFDRTAGLFHSILYTKL
FSVLLLALSCLGTKGVKGEKITWGRIWTAFAVGFVLFFLNWWLLPLPLPLEAVTGLYVLTIGTGYVCLLM
GGLWMSRLLKHNLMEDVFNNENESFMQETRLIESEYSVNLPTRFYYRKRWNNGWINVVNPFRASIVLGTP
GSGKSYAVVNNFIKQQIEKGFSQYIYDFKYPDLSTIAYNHLLNHPDGYKVKPKFYVINFDDPRRSHRCNP
IHPDFMEDITDAYESAYTIMLNLNKTWVQKQGDFFVESPIILFASIIWYLKIYQNGKFCTFPHAIEFLNR
RYEDIFPILTSYPELENYLSPFMDAWLGGAAEQLMGQIASAKIPLSRMISPQLYWVMSDSEFTLDINNPE
EPKILCVGNNPDRQNIYGAALGLYNSRIVKLINKKGMLKSSVIIDELPTIYFKGLDNLIATARSNKVAVC
LGFQDFSQLVRDYGDKEAKVVMNTVGNIFSGQVVGETAKTLSERFGKVLQKRQSISINRQDVSTSINTQM
DALIPPSKISGLTQGMFVGSVSDNFNERIEQKIFHCEIVVDAEKVKREESAYKKIPVITNFTDEDGNDRM
KETVQANYRRIKEEVKQIVQEELERIKNDPVLCKLLPDNETV
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q56VF1 |
Reference
[1] Lindsay Peed et al. (2010) Genetic and functional analyses of the mob operon on conjugative transposon CTn341 from Bacteroides spp. Journal of bacteriology. 192(18):4643-50. [PMID:20639338]
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 5709..16293
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_0 | 1056..1451 | + | 396 | AAS83474 | Ctn001 | - |
| Locus_1 | 1465..1893 | + | 429 | AAS83475 | Ctn002 | - |
| Locus_2 | 1890..3173 | + | 1284 | AAS83476 | Ctn003 | - |
| Locus_3 | 3702..3950 | + | 249 | AAS83477 | Ctn004 | - |
| Locus_4 | 4004..4312 | + | 309 | AAS83478 | YahA | - |
| Locus_5 | 4477..5112 | + | 636 | AAS83479 | Ctn006 | - |
| Locus_6 | 5185..5712 | - | 528 | AAS83480 | Lys | - |
| Locus_7 | 5709..6215 | - | 507 | AAS83481 | TraQ | traQ |
| Locus_8 | 6234..7112 | - | 879 | AAS83482 | TraP | traP |
| Locus_9 | 7121..7696 | - | 576 | AAS83483 | TraO | traO |
| Locus_10 | 7699..8685 | - | 987 | AAS83484 | TraN | traN |
| Locus_11 | 8754..10103 | - | 1350 | AAS83485 | TraM | traM |
| Locus_12 | 10084..10392 | - | 309 | AAS83486 | TraL | traL |
| Locus_13 | 10398..11021 | - | 624 | AAS83487 | TraK | traK |
| Locus_14 | 11053..12057 | - | 1005 | AAS83488 | TraJ | traJ |
| Locus_15 | 12061..12690 | - | 630 | AAS83489 | TraI | traI |
| Locus_16 | 12717..13106 | - | 390 | AAS83490 | TraH | traH |
| Locus_17 | 13138..15822 | - | 2685 | AAS83491 | TraG | virb4 |
| Locus_18 | 15639..15971 | - | 333 | AAS83492 | TraF | traF |
| Locus_19 | 15982..16293 | - | 312 | AAS83493 | TraE | traE |
| Locus_20 | 16494..17234 | - | 741 | AAS83494 | TraD | - |
| Locus_21 | 17231..17593 | - | 363 | AAS83495 | TraC | - |
| Locus_22 | 17617..18066 | - | 450 | AAS83496 | TraB | - |
| Locus_23 | 18072..18827 | - | 756 | AAS83497 | TraA | - |
| Locus_24 | 19509..19937 | + | 429 | AAS83498 | MobA | - |
| Locus_25 | 19916..21163 | + | 1248 | AAS83499 | MobB | - |
Host bacterium
| ID | 102 | Element type | ICE (Integrative and conjugative element) |
| Element name | CTn341 | GenBank | AY515263 |
| Element size | 51993 bp | Coordinate of oriT [Strand] | 19315..19514 [+] |
| Host bacterium | Bacteroides fragilis |
Cargo genes
| Drug resistance gene | tet(Q) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9 |