Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200092 |
| Name | oriT_Trb-1 |
| Organism | Legionella pneumophila str. Corby |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | CP000675 (637526..637939 [+], 414 nt) |
| oriT length | 414 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note |
oriT sequence
Download Length: 414 nt
AAGGATGCTGACCTCTTAACAAGTGGTTTGCAATCCTTTAGGTACTGATTAATTGTTTTCTTTTTGATCTCAAAAAGCAAGATACCCGTTGAGCACCGAAGGTGCGAATAAGGTTTGTGAAGATTGATAGCCAGCTTGCTGGTTAGCTAACTTCACCTATCTTGCCCTGCTTTTCCAGATCACTTTAAAGAATCATATCAGGAACTTTTTGGAACTTGCAACGATTTTTAAAGAACTTTTTTGCCTATAGAACTAAAATAATCCAAATTATTCCTAAAAGTTCTTTTATAATGCTCAAATACTCATTTTTATATTCTAGTAAATTCTTAATAGTTCTAAAATATTCCTTTTTTCTTGTTCCAATCCTAATTATGATTTAGACTTTTTCAATAATAAAATTGACACCAAAGAAGA
oriT secondary structure
Predicted by RNAfold.
Download structure file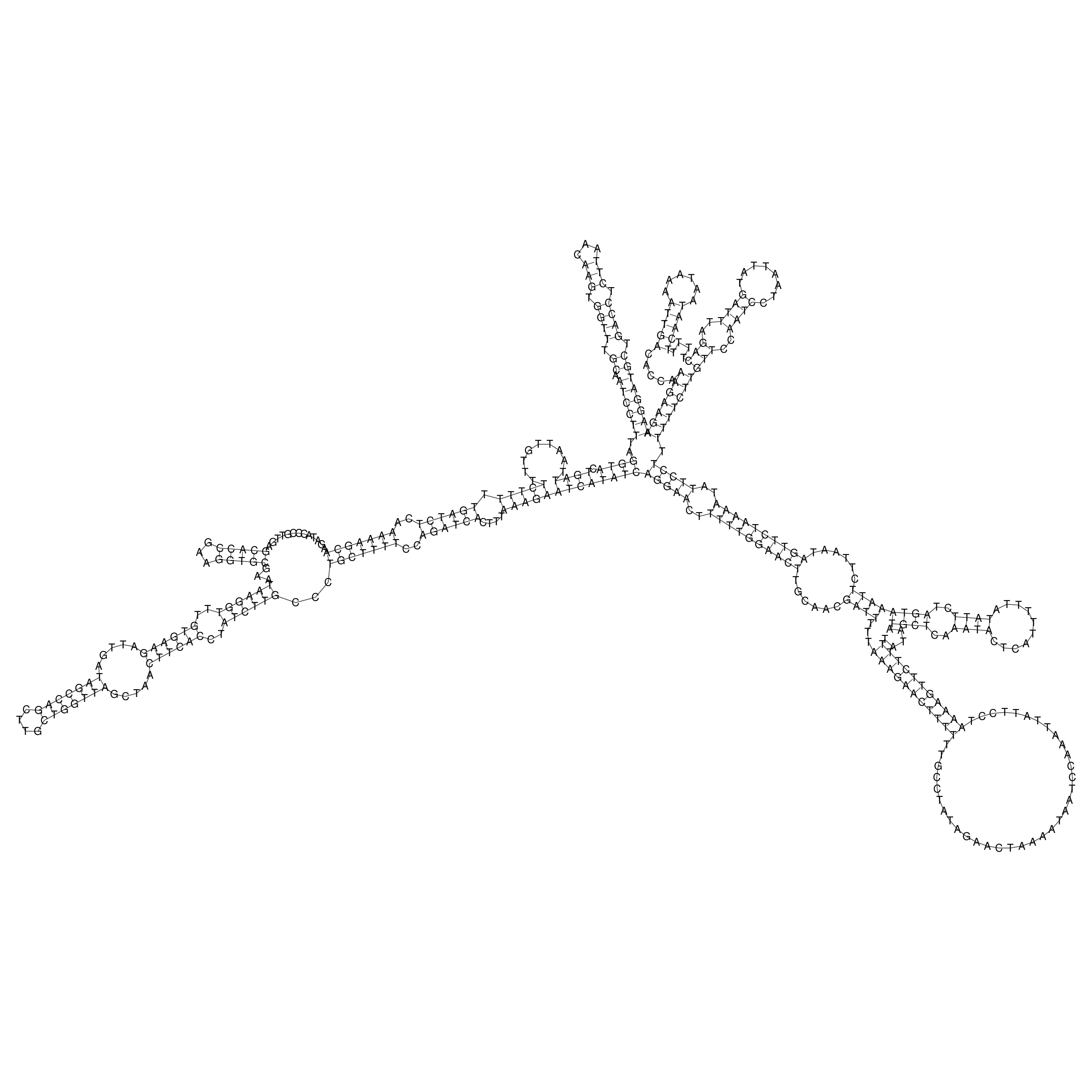
Reference
[1] Gernot Glöckner et al. (2008) Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. International journal of medical microbiology : IJMM. 298(5-6):411-28. [PMID:17888731]
Relaxase
| ID | 1096 | GenBank | ABQ56696 |
| Name | traI_Trb-1 |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | |
| Note | |||
Relaxase protein sequence
Download Length: 621 a.a. Molecular weight: 71627.04 Da Isoelectric Point: 10.5877
MIIRHIPMKSARLSSFSALVKYITDEQNKQERVGKVRISNCNSVDPTWAIHEVLATQARNQRAKSDKTYH
MLISFAPGENPPNEVLKAIEERIISSIGFKDHQRISAIHHDTDNLHIHVAINKIHPKAFNIIEPYRAYRT
FSEVASKLEIEYGLEITNHQTRKSRSENLADDMEQHSGIESLINWIKRNCLEQINQANHWNEVHKILSLH
GLEIHIKANGLVFYNEAGLMVKASSVSRCFSKKNLESRLGEFKPSPFSTERSRQNIYRYEPLNRKVIGSE
IYARYLYEKEHGKTLLAEKLKHLREAKARLITKAKKRGRTKRAVLKLMKAPRTQKKYLYQQISKTLLKEI
EKVRQNYAKERKHLLELHKNKTWADWLQQKAREGDKEALTAMRYHNRKNQSNYSLSAASSNFSLADIKQV
DSITKEGTEIYKMDKAVIRNTGKEIKISKGGSIATLKKAIEMAQQRYGNCIQVNGSPLFKKIILQITIQH
NIPLTFADSEMEAQRQKMVLEQERRYEQSRRYRFNDGGRTPRSNEAAGAAIGERGTLSSTKPNADSIRQG
PPAKSQNSLRDLSQLDMVQLARRSKVLLPDNAHDQLERQRLKSDNYVRRKVSGLSRKVEHK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Reference
[1] Gernot Glöckner et al. (2008) Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. International journal of medical microbiology : IJMM. 298(5-6):411-28. [PMID:17888731]
Auxiliary protein
| ID | 392 | GenBank | ABQ56695 |
| Name | traJ1_Trb-1 |
UniProt ID | _ |
| Length | 117 a.a. | PDB ID | _ |
| Note | |||
Auxiliary protein sequence
Download Length: 117 a.a. Molecular weight: 13328.57 Da Isoelectric Point: 10.7747
MDDKNKSPSRKHGRHLRVPVLPDEEISIKSHAAQAGLSVAGYLRRIGLGYQIHSAIDKDYILQLSKINAD
MGRLGGLFKLWLTQDRRVAHFDHRKVKALLDRIQTMQAAMFEVVKKL
Protein domains
No domain identified.
Protein structure
No available structure.
Reference
[1] Gernot Glöckner et al. (2008) Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. International journal of medical microbiology : IJMM. 298(5-6):411-28. [PMID:17888731]
| ID | 393 | GenBank | ABQ56694 |
| Name | traK_Trb-1 |
UniProt ID | _ |
| Length | 108 a.a. | PDB ID | _ |
| Note | |||
Auxiliary protein sequence
Download Length: 108 a.a. Molecular weight: 12488.28 Da Isoelectric Point: 9.8029
MKKPLSERVIQNQSEKKNGRTNAKIQFIALKEDIREALDKGCSMKAVWETLSDEGQISFGYKAFRHYVLK
LIKSEQENIRDDKQSKSKTTNEIKGFTFNPIPNPEELL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Reference
[1] Gernot Glöckner et al. (2008) Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. International journal of medical microbiology : IJMM. 298(5-6):411-28. [PMID:17888731]
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 619346..629196
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LPC_2819 | 614497..615231 | + | 735 | ABQ56720 | hypothetical protein | - |
| LPC_2818 | 615234..616457 | + | 1224 | ABQ56719 | site specific recombinase | - |
| LPC_2817 | 616462..616881 | + | 420 | ABQ56718 | hypothetical protein | - |
| LPC_2816 | 616983..617657 | - | 675 | ABQ56717 | hypothetical protein | - |
| LPC_2815 | 617842..618723 | + | 882 | ABQ56716 | Legionella vir region protein | - |
| LPC_2814 | 618740..619048 | + | 309 | ABQ56715 | Legionella vir region protein | - |
| LPC_2813 | 619069..619335 | + | 267 | ABQ56714 | global regulator (carbon storage regulator) | - |
| LPC_2812 | 619346..620311 | + | 966 | ABQ56713 | LvhB11 | virB11 |
| LPC_2811 | 620323..620700 | + | 378 | ABQ56712 | hypothetical protein | virB2 |
| LPC_2810 | 620751..620996 | + | 246 | ABQ56711 | Conjugal transfer protein trbD | virB3 |
| LPC_2809 | 620993..623518 | + | 2526 | ABQ56710 | Conjugal transfer protein trbE precursor | virb4 |
| LPC_2808 | 623515..624258 | + | 744 | ABQ56709 | Conjugal transfer protein trbF | virB8 |
| LPC_2807 | 624255..625130 | + | 876 | ABQ56708 | Conjugal transfer protein trbG precursor | virB9 |
| LPC_2806 | 625133..625543 | + | 411 | ABQ56707 | hypothetical protein | - |
| LPC_2805 | 625549..626799 | + | 1251 | ABQ56706 | Conjugal transfer protein trbI | virB10 |
| LPC_2804 | 626815..627555 | + | 741 | ABQ56705 | Conjugal transfer protein trbJ precursor | virB5 |
| LPC_2803 | 627582..627794 | + | 213 | ABQ56704 | hypothetical protein | - |
| LPC_2802 | 627799..629196 | + | 1398 | ABQ56703 | Conjugal transfer protein trbL | virB6 |
| LPC_2801 | 629193..631082 | + | 1890 | ABQ56702 | Conjugal transfer protein traG | - |
| LPC_2800 | 631079..631624 | + | 546 | ABQ56701 | Plasmid transfer protein traF precursor | - |
| LPC_2799 | 631621..631785 | + | 165 | ABQ56700 | traD protein | - |
| LPC_2798 | 631778..633964 | + | 2187 | ABQ56699 | DNA primase traC (Replication primase) | - |
Region 2: 185810..648783
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LPC_0163 | 181312..181422 | + | 111 | ABQ54162 | hypothetical protein | - |
| LPC_0164 | 181809..181970 | + | 162 | ABQ54163 | hypothetical protein | - |
| LPC_0165 | 182031..183335 | - | 1305 | ABQ54164 | hypothetical protein | - |
| LPC_0166 | 183470..184129 | - | 660 | ABQ54165 | phage repressor | - |
| LPC_0167 | 184300..185184 | + | 885 | ABQ54166 | Legionella vir region protein | - |
| LPC_0168 | 185201..185512 | + | 312 | ABQ54167 | Legionella vir region protein | - |
| LPC_0169 | 185531..185797 | + | 267 | ABQ54168 | global regulator (carbon storage regulator) | - |
| LPC_0170 | 185810..186775 | + | 966 | ABQ54169 | LvhB11 | virB11 |
| LPC_0171 | 186788..187165 | + | 378 | ABQ54170 | Conjugal transfer protein trbC | virB2 |
| LPC_0172 | 187162..187461 | + | 300 | ABQ54171 | Conjugal transfer protein trbD | virB3 |
| LPC_0173 | 187458..189995 | + | 2538 | ABQ54172 | Conjugal transfer protein trbE precursor | virb4 |
| LPC_0174 | 189992..190735 | + | 744 | ABQ54173 | Conjugal transfer protein trbF | virB8 |
| LPC_0175 | 190732..191607 | + | 876 | ABQ54174 | conjugal transfer protein trbG precursor | virB9 |
| LPC_0176 | 191610..192020 | + | 411 | ABQ54175 | hypothetical protein | - |
| LPC_0177 | 192026..193267 | + | 1242 | ABQ54176 | Conjugal transfer protein trbI | virB10 |
| LPC_0178 | 193281..194021 | + | 741 | ABQ54177 | Conjugal transfer protein trbJ precursor | virB5 |
| LPC_0179 | 194051..194263 | + | 213 | ABQ54178 | hypothetical protein | - |
| LPC_0180 | 194211..195683 | + | 1473 | ABQ54179 | Probable conjugal transfer protein trbL | virB6 |
| LPC_0181 | 195680..197569 | + | 1890 | ABQ54180 | Conjugal transfer protein traG | - |
| LPC_0182 | 197566..198111 | + | 546 | ABQ54181 | Conjugal transfer protein traF precursor | - |
| LPC_0183 | 198108..198272 | + | 165 | ABQ54182 | Protein traD | - |
| LPC_0184 | 198265..200451 | + | 2187 | ABQ54183 | DNA primase traC (Replication primase) | - |
| LPC_0185 | 200533..200904 | - | 372 | ABQ54184 | hypothetical protein | - |
| LPC_0186 | 200882..201940 | - | 1059 | ABQ54185 | hypothetical protein | - |
| LPC_0187 | 202055..203929 | - | 1875 | ABQ54186 | TraI protein | - |
| LPC_0188 | 203926..204279 | - | 354 | ABQ54187 | TraJ protein (Relaxosome protein) | - |
| LPC_0190 | 207613..208338 | + | 726 | ABQ54188 | hypothetical protein | - |
| LPC_0191 | 208338..208793 | + | 456 | ABQ54189 | hypothetical protein | - |
| LPC_0192 | 208807..209451 | + | 645 | ABQ54190 | hypothetical protein | - |
| LPC_0193 | 209495..209956 | - | 462 | ABQ54191 | cytidine/deoxycytidylate deaminase | - |
| LPC_0194 | 209934..211376 | - | 1443 | ABQ54192 | hypothetical protein | - |
| LPC_0195 | 211369..211728 | - | 360 | ABQ54193 | hypothetical protein | - |
| LPC_0196 | 211746..212399 | - | 654 | ABQ54194 | hypothetical protein | - |
| LPC_0197 | 212528..212776 | + | 249 | ABQ54195 | hypothetical protein | - |
| LPC_0198 | 212810..213016 | + | 207 | ABQ54196 | hypothetical protein | - |
| LPC_0199 | 213091..214170 | + | 1080 | ABQ54197 | Putative lambdoid prophage Rac integrase | - |
| LPC_0200 | 214176..214991 | - | 816 | ABQ54198 | hypothetical protein | - |
| LPC_0201 | 214988..215779 | - | 792 | ABQ54199 | hypothetical protein | - |
| LPC_0202 | 216356..217576 | + | 1221 | ABQ54200 | Integrase | - |
| LPC_0203 | 217598..218434 | + | 837 | ABQ54201 | DNA-damage-inducible protein D | - |
| LPC_0204 | 218455..219234 | + | 780 | ABQ54202 | hypothetical protein | - |
| LPC_0205 | 219385..221175 | - | 1791 | ABQ54203 | Probable protease htpX like protein | - |
| LPC_0206 | 221543..221800 | - | 258 | ABQ54204 | hypothetical protein | - |
| LPC_0207 | 221942..222466 | + | 525 | ABQ54205 | hypothetical protein | - |
| LPC_0208 | 222604..222804 | + | 201 | ABQ54206 | Prophage CP4-57 regulatory protein alpA | - |
| LPC_0209 | 222820..222945 | + | 126 | ABQ54207 | hypothetical protein | - |
| LPC_0210 | 222938..225343 | + | 2406 | ABQ54208 | 5' DNA primase traC (Replication primase) | - |
| LPC_0211 | 225333..225644 | + | 312 | ABQ54209 | hypothetical protein | - |
| LPC_0212 | 225704..226078 | + | 375 | ABQ54210 | hypothetical protein | - |
| LPC_0213 | 226092..226850 | - | 759 | ABQ54211 | hypothetical protein | - |
| LPC_0214 | 226936..227196 | + | 261 | ABQ54212 | hypothetical protein | - |
| LPC_0215 | 227257..229413 | + | 2157 | ABQ54213 | hypothetical protein | - |
| LPC_0216 | 229443..230015 | - | 573 | ABQ54214 | hypothetical protein | - |
| LPC_0218 | 232516..233283 | - | 768 | ABQ54215 | hypothetical protein | - |
| LPC_0219 | 233982..234959 | + | 978 | ABQ54216 | Mrr restriction system protein (EcoKMrr) | - |
| LPC_0220 | 235158..235505 | + | 348 | ABQ54217 | hypothetical protein | - |
| LPC_0221 | 235601..235786 | + | 186 | ABQ54218 | hypothetical protein | - |
| LPC_0222 | 235776..236246 | + | 471 | ABQ54219 | Putative endonuclease precursor | - |
| LPC_0223 | 236835..237590 | + | 756 | ABQ54220 | hypothetical protein | - |
| LPC_0224 | 237637..237777 | + | 141 | ABQ54221 | hypothetical protein | - |
| LPC_0225 | 238077..240572 | + | 2496 | ABQ54222 | hypothetical protein | - |
| LPC_0226 | 240854..240946 | - | 93 | ABQ54223 | hypothetical protein | - |
| LPC_0227 | 240970..241161 | - | 192 | ABQ54224 | hypothetical protein | - |
| LPC_0228 | 241325..241840 | + | 516 | ABQ54225 | hypothetical protein | - |
| LPC_0229 | 242290..242514 | - | 225 | ABQ54226 | hypothetical protein | - |
| LPC_0230 | 242731..243045 | - | 315 | ABQ54227 | hypothetical protein | - |
| LPC_0231 | 243449..244885 | + | 1437 | ABQ54228 | hypothetical protein | - |
| LPC_0232 | 245146..245892 | + | 747 | ABQ54229 | hypothetical protein | - |
| LPC_0233 | 246366..247076 | + | 711 | ABQ54230 | ABC transporter, ATP binding protein | - |
| LPC_0234 | 247070..249436 | + | 2367 | ABQ54231 | hypothetical protein | - |
| LPC_0235 | 249436..250635 | + | 1200 | ABQ54232 | hypothetical protein | - |
| LPC_0236 | 250707..251585 | - | 879 | ABQ54233 | sensory box (GGDEF/EAL domain) | - |
| LPC_0237 | 251569..252021 | - | 453 | ABQ54234 | hypothetical protein | - |
| LPC_0238 | 252126..252887 | - | 762 | ABQ54235 | methionine aminopeptidase | - |
| LPC_0239 | 252868..253080 | - | 213 | ABQ54236 | hypothetical protein | - |
| LPC_0240 | 253256..253735 | - | 480 | ABQ54237 | hypothetical protein | - |
| LPC_0241 | 253748..253987 | - | 240 | ABQ54238 | hypothetical protein | - |
| LPC_0242 | 254047..255024 | + | 978 | ABQ54239 | hypothetical protein | - |
| LPC_0243 | 255117..255854 | + | 738 | ABQ54240 | hypothetical protein | - |
| LPC_0244 | 255992..256450 | - | 459 | ABQ54241 | hypothetical protein | - |
| LPC_0245 | 256837..257610 | + | 774 | ABQ54242 | hydrolases of the alpha/beta superfamily | - |
| LPC_0246 | 257692..258147 | - | 456 | ABQ54243 | hypothetical protein | - |
| LPC_0247 | 258316..259200 | - | 885 | ABQ54244 | hypothetical protein | - |
| LPC_0248 | 259476..259886 | - | 411 | ABQ54245 | peptide chain release factor | - |
| LPC_0249 | 259987..260697 | - | 711 | ABQ54246 | conserved hypothetical protein containing DUF330 domain protein | - |
| LPC_0250 | 260694..261380 | - | 687 | ABQ54247 | probable transmembrane protein; conserved hypothetical protein | - |
| LPC_0251 | 261448..262203 | - | 756 | ABQ54248 | probable ABC transport system permease protein | - |
| LPC_0252 | 262507..262650 | - | 144 | ABQ54249 | hypothetical protein | - |
| LPC_0253 | 262869..263567 | + | 699 | ABQ54250 | hypothetical protein | - |
| LPC_0255 | 263692..264552 | - | 861 | ABQ54251 | transcriptional regulator, LysR | - |
| LPC_0256 | 264663..265712 | + | 1050 | ABQ54252 | pyoverdine biosynthesis protein PvcA | - |
| LPC_0257 | 265702..266538 | + | 837 | ABQ54253 | pyoverdine biosynthesis protein PvcB | - |
| LPC_0258 | 266535..267977 | + | 1443 | ABQ54254 | FAD monooxygenase, PheA/TfdB family | - |
| LPC_0259 | 267974..269122 | + | 1149 | ABQ54255 | chloramphenicol resistance protein | - |
| LPC_0260 | 269119..270351 | + | 1233 | ABQ54256 | hypothetical protein | - |
| LPC_0261 | 270519..271661 | - | 1143 | ABQ54257 | hypothetical protein | - |
| LPC_0262 | 271827..271988 | - | 162 | ABQ54258 | hypothetical protein | - |
| LPC_0263 | 272210..273184 | + | 975 | ABQ54259 | O-methyltransferase | - |
| LPC_0264 | 273310..274152 | - | 843 | ABQ54260 | heat shock protein, protease HtpX | - |
| LPC_0265 | 274273..275265 | + | 993 | ABQ54261 | hypothetical protein | - |
| LPC_0266 | 275470..276948 | + | 1479 | ABQ54262 | amine oxidase, flavin containing | - |
| LPC_0267 | 277208..278617 | + | 1410 | ABQ54263 | zinc metalloprotein | - |
| LPC_0268 | 278756..279808 | + | 1053 | ABQ54264 | acyl CoA transferase/carnitine dehydratase | - |
| LPC_0269 | 280005..280871 | - | 867 | ABQ54265 | hypothetical protein | - |
| LPC_0270 | 281537..281935 | - | 399 | ABQ54266 | heat shock hsp20 | - |
| LPC_0271 | 282419..284668 | - | 2250 | ABQ54267 | catalase/(hydro)peroxidase KatG | - |
| LPC_0272 | 284799..285791 | - | 993 | ABQ54268 | hypothetical protein | - |
| LPC_0273 | 286272..286589 | + | 318 | ABQ54269 | hypothetical protein | - |
| LPC_0274 | 286787..288157 | + | 1371 | ABQ54270 | cytochrome D ubiquinol oxidase subunit I | - |
| LPC_0275 | 288157..289146 | + | 990 | ABQ54271 | hypothetical protein | - |
| LPC_0277 | 289147..289923 | - | 777 | ABQ54272 | hypothetical protein | - |
| LPC_0278 | 289929..290465 | - | 537 | ABQ54273 | hypothetical protein | - |
| LPC_0279 | 290455..292242 | - | 1788 | ABQ54274 | fusion of two types of conserved hypothetical protein | - |
| LPC_0280 | 292325..293023 | + | 699 | ABQ54275 | 2-deoxy-D-gluconate-3-dehydrogenase | - |
| LPC_0281 | 293040..294542 | + | 1503 | ABQ54276 | hypothetical protein | - |
| LPC_0282 | 294583..294996 | - | 414 | ABQ54277 | membrane protein | - |
| LPC_0283 | 295371..298256 | + | 2886 | ABQ54278 | serine/threonine-protein kinase | - |
| LPC_0284 | 298275..300245 | + | 1971 | ABQ54279 | hypothetical protein | - |
| LPC_0285 | 300326..300913 | + | 588 | ABQ54280 | hypothetical protein | - |
| LPC_0286 | 300918..301046 | - | 129 | ABQ54281 | hypothetical protein | - |
| LPC_0287 | 301079..302494 | - | 1416 | ABQ54282 | deoxyribodipyrimidine photolyase | - |
| LPC_0288 | 302590..303297 | - | 708 | ABQ54283 | inner membrane protein, LrgB family protein | - |
| LPC_0289 | 303275..303664 | - | 390 | ABQ54284 | murein hydrolase exporter, LrgA family protein | - |
| LPC_0290 | 303770..304654 | + | 885 | ABQ54285 | transcriptional regulator, LysR family | - |
| LPC_0291 | 304641..304751 | + | 111 | ABQ54286 | hypothetical protein | - |
| LPC_0292 | 304776..304988 | + | 213 | ABQ54287 | conserved hypothetical protein; DUF329 | - |
| LPC_0293 | 305074..306153 | - | 1080 | ABQ54288 | phosphoribosylaminoimidazole carboxylase, ATPase subunit | - |
| LPC_0294 | 306150..306650 | - | 501 | ABQ54289 | phosphoribosylaminoimidazole carboxylase, catalytic subunit PurE | - |
| LPC_0295 | 306710..307549 | - | 840 | ABQ54290 | hypothetical protein | - |
| LPC_0296 | 307574..307927 | - | 354 | ABQ54291 | hypothetical protein | - |
| LPC_0297 | 307941..310229 | - | 2289 | ABQ54292 | hypothetical protein | - |
| LPC_0298 | 310244..311764 | - | 1521 | ABQ54293 | NADH dehydrogenase subunit 5 | - |
| LPC_0299 | 311866..312717 | + | 852 | ABQ54294 | transcriptional regulator, LysR family | - |
| LPC_0300 | 312759..314348 | + | 1590 | ABQ54295 | toxin secretion ABC transporter HlyB/MsbA family ATP-binding protein | - |
| LPC_0301 | 314345..315331 | + | 987 | ABQ54296 | RND efflux membrane fusion protein | - |
| LPC_0302 | 315328..316725 | + | 1398 | ABQ54297 | multidrug efflux protein, outer membrane component | - |
| LPC_0303 | 316987..318009 | + | 1023 | ABQ54298 | hypothetical protein | - |
| LPC_0304 | 318424..319257 | - | 834 | ABQ54299 | heme oxygenase | - |
| LPC_0305 | 319469..321613 | - | 2145 | ABQ54300 | sensor histidine kinase | - |
| LPC_0306 | 321758..324316 | - | 2559 | ABQ54301 | heavy metal transporting P-type ATPase, cation transporting | - |
| LPC_0307 | 324561..325094 | + | 534 | ABQ54302 | transcriptional regulator np20, Fur family | - |
| LPC_0308 | 325212..326804 | - | 1593 | ABQ54303 | benzoylformate decarboxylase | - |
| LPC_0309 | 327100..331590 | - | 4491 | ABQ54304 | SidE protein, substrate of the Dot/Icm system | - |
| LPC_0310 | 331845..332024 | - | 180 | ABQ54305 | hypothetical protein | - |
| LPC_0311 | 332014..332496 | - | 483 | ABQ54306 | hypothetical protein | - |
| LPC_0312 | 332590..334569 | - | 1980 | ABQ54307 | hypothetical protein | - |
| LPC_0313 | 334851..335645 | + | 795 | ABQ54308 | lipolytic enzyme | - |
| LPC_0314 | 335661..337127 | + | 1467 | ABQ54309 | glycine betaine aldehyde dehydrogenase | - |
| LPC_0315 | 337132..338454 | + | 1323 | ABQ54310 | 4-aminobutyrate aminotransferase | - |
| LPC_0316 | 338543..339310 | + | 768 | ABQ54311 | DNA repair protein | - |
| LPC_0317 | 339289..339420 | - | 132 | ABQ54312 | hypothetical protein | - |
| LPC_0318 | 339669..340478 | + | 810 | ABQ54313 | hypothetical protein | - |
| LPC_0319 | 340620..341552 | + | 933 | ABQ54314 | glutaminase | - |
| LPC_0320 | 341654..342253 | - | 600 | ABQ54315 | short chain dehydrogenase | - |
| LPC_0321 | 342584..343978 | - | 1395 | ABQ54316 | pyridine nucleotide-disulfide oxidoreductase | - |
| LPC_0322 | 344030..347392 | - | 3363 | ABQ54317 | NAD-glutamate dehydrogenase | - |
| LPC_0323 | 347742..348314 | + | 573 | ABQ54318 | hypothetical protein | - |
| LPC_0324 | 348554..349507 | + | 954 | ABQ54319 | hypothetical protein | - |
| LPC_0325 | 349583..350008 | + | 426 | ABQ54320 | arsenate reductase | - |
| LPC_0326 | 350094..351245 | - | 1152 | ABQ54321 | stearoyl CoA 9-desaturase | - |
| LPC_0327 | 351367..352611 | - | 1245 | ABQ54322 | ATP-dependent RNA helicase, DEAD box family | - |
| LPC_0328 | 352942..353217 | + | 276 | ABQ54323 | RNA binding protein, cold-inducible rrm | - |
| LPC_0329 | 353394..354071 | + | 678 | ABQ54324 | membrane protein | - |
| LPC_0330 | 354109..354747 | + | 639 | ABQ54325 | acetyltransferase, GNAT family | - |
| LPC_0331 | 354882..355409 | + | 528 | ABQ54326 | hypothetical protein | - |
| LPC_0332 | 355564..356910 | + | 1347 | ABQ54327 | outer membrane channel protein | - |
| LPC_0333 | 356900..357937 | + | 1038 | ABQ54328 | conserved domain protein | - |
| LPC_0334 | 357994..358986 | + | 993 | ABQ54329 | multidrug resistance secretion protein | - |
| LPC_0335 | 359162..361105 | + | 1944 | ABQ54330 | hypothetical protein | - |
| LPC_0336 | 361124..362908 | + | 1785 | ABQ54331 | hypothetical protein | - |
| LPC_0337 | 363157..363477 | + | 321 | ABQ54332 | hypothetical protein | - |
| LPC_0338 | 363563..364201 | - | 639 | ABQ54333 | bacteriophage related DNA polymerase | - |
| LPC_0339 | 364458..365732 | + | 1275 | ABQ54334 | MFS transporter family protein | - |
| LPC_0340 | 365811..366509 | + | 699 | ABQ54335 | N-acetylmuramoyl-L-alanine amidase; amidase 2 | - |
| LPC_0341 | 366564..368105 | - | 1542 | ABQ54336 | hypothetical protein | - |
| LPC_0342 | 368334..368768 | + | 435 | ABQ54337 | hypothetical protein | - |
| LPC_0343 | 368783..369763 | - | 981 | ABQ54338 | hypothetical protein | - |
| LPC_0344 | 370042..370638 | + | 597 | ABQ54339 | hypothetical protein | - |
| LPC_0345 | 370773..372272 | + | 1500 | ABQ54340 | hypothetical protein | - |
| LPC_0346 | 372353..373756 | + | 1404 | ABQ54341 | nicotinate phosphoribosyltransferase | - |
| LPC_0347 | 373761..374381 | + | 621 | ABQ54342 | bifunctional pyrazinamidase/nicotinamidase | - |
| LPC_0348 | 374463..374846 | - | 384 | ABQ54343 | cysteine transferase | - |
| LPC_0349 | 374846..376060 | - | 1215 | ABQ54344 | major facilitator superfamily transporter | - |
| LPC_0350 | 376178..377041 | + | 864 | ABQ54345 | transcriptional regulator, LysR family | - |
| LPC_0351 | 377462..378172 | + | 711 | ABQ54346 | hypothetical protein | - |
| LPC_0352 | 378239..380812 | + | 2574 | ABQ54347 | SdbA protein, putative substrate of the Dot/Icm system | - |
| LPC_0353 | 380892..382445 | + | 1554 | ABQ54348 | hypothetical protein | - |
| LPC_0354 | 382511..384736 | - | 2226 | ABQ54349 | sensory box protein, GGDEF family protein, LssE | - |
| LPC_0355 | 384739..386154 | - | 1416 | ABQ54350 | sensor histidine kinase | - |
| LPC_0356 | 386179..387345 | - | 1167 | ABQ54351 | hypothetical protein | - |
| LPC_0357 | 387515..388396 | - | 882 | ABQ54352 | transcriptional regulator, LysR family | - |
| LPC_0358 | 388618..389802 | - | 1185 | ABQ54353 | amino acid transporter | - |
| LPC_0359 | 389870..390646 | - | 777 | ABQ54354 | hypothetical protein | - |
| LPC_0360 | 390854..392065 | + | 1212 | ABQ54355 | NAD dependent formate dehydrogenase | - |
| LPC_0361 | 392171..393295 | - | 1125 | ABQ54356 | hypothetical protein | - |
| LPC_0362 | 393552..394214 | + | 663 | ABQ54357 | hypothetical protein | - |
| LPC_0363 | 394228..394341 | + | 114 | ABQ54358 | hypothetical protein | - |
| LPC_0364 | 394322..395095 | + | 774 | ABQ54359 | oxidoreductase, short chain dehydrogenase/reductase family | - |
| LPC_0365 | 395614..395844 | + | 231 | ABQ54360 | hypothetical protein | - |
| LPC_0366 | 395898..396467 | - | 570 | ABQ54361 | hypothetical protein | - |
| LPC_0367 | 396539..397519 | + | 981 | ABQ54362 | hypothetical protein | - |
| LPC_0368 | 397588..399660 | - | 2073 | ABQ54363 | polyphosphate kinase | - |
| LPC_0369 | 399701..400906 | - | 1206 | ABQ54364 | lipoprotein | - |
| LPC_0370 | 400982..401515 | - | 534 | ABQ54365 | chromate transport protein | - |
| LPC_0371 | 401512..402132 | - | 621 | ABQ54366 | hypothetical protein conserved within Legionellae | - |
| LPC_0372 | 402308..404746 | + | 2439 | ABQ54367 | long chain acyl-CoA dehydrogenase | - |
| LPC_0373 | 404860..405552 | + | 693 | ABQ54368 | hypothetical protein | - |
| LPC_0374 | 405589..406251 | - | 663 | ABQ54369 | mannose-1-phosphate guanyltransferase | - |
| LPC_0375 | 406248..407225 | - | 978 | ABQ54370 | hypothetical phosphotransferase | - |
| LPC_0376 | 407333..409987 | + | 2655 | ABQ54371 | organic solvent tolerance protein | - |
| LPC_0377 | 410163..411452 | + | 1290 | ABQ54372 | peptidyl-prolyl cis-trans isomerase D (SurA) | - |
| LPC_0378 | 411449..412423 | + | 975 | ABQ54373 | 4-hydroxythreonine-4-phosphate dehydrogenase PdxA | - |
| LPC_0379 | 412425..412916 | + | 492 | ABQ54374 | dihydrofolate reductase | - |
| LPC_0380 | 412923..413468 | - | 546 | ABQ54375 | hypothetical protein | - |
| LPC_3028 | 419343..420533 | + | 1191 | ABQ56927 | translation elongation factor Tu (EF-Tu); tRNA- Ala | - |
| LPC_3026 | 421058..421606 | + | 549 | ABQ56926 | transcription antitermination protein NusG | - |
| LPC_3025 | 421716..422150 | + | 435 | ABQ56925 | hypothetical protein | - |
| LPC_3024 | 422160..422855 | + | 696 | ABQ56924 | hypothetical protein | - |
| LPC_3023 | 423028..423561 | + | 534 | ABQ56923 | hypothetical protein | - |
| LPC_3022 | 423592..423972 | + | 381 | ABQ56922 | 50S ribosomal protein L7/L12 | - |
| LPC_3021 | 424064..428170 | + | 4107 | ABQ56921 | DNA-directed RNA polymerase beta subunit | - |
| LPC_3020 | 428241..432464 | + | 4224 | ABQ56920 | hypothetical protein | - |
| LPC_3019 | 432578..432958 | + | 381 | ABQ56919 | hypothetical protein | - |
| LPC_3018 | 432979..433506 | + | 528 | ABQ56918 | hypothetical protein | - |
| LPC_3017 | 433521..435605 | + | 2085 | ABQ56917 | translation elongation factor G (EF-G) | - |
| LPC_3015 | 435626..436816 | + | 1191 | ABQ56916 | translation elongation factor Tu (EF-Tu) | - |
| LPC_3014 | 436822..437139 | + | 318 | ABQ56915 | 30S ribosomal protein S10 | - |
| LPC_3013 | 437174..437824 | + | 651 | ABQ56914 | 50S ribosomal protein L3 | - |
| LPC_3012 | 437824..438429 | + | 606 | ABQ56913 | 50S ribosomal protein L4 | - |
| LPC_3011 | 438426..438722 | + | 297 | ABQ56912 | 50S ribosomal protein L23 | - |
| LPC_3010 | 438734..439561 | + | 828 | ABQ56911 | 50S ribosomal protein L2 | - |
| LPC_3009 | 439580..439858 | + | 279 | ABQ56910 | 30S ribosomal subunit protein S19 | - |
| LPC_3008 | 439868..440203 | + | 336 | ABQ56909 | 50S ribosomal protein L22 | - |
| LPC_3007 | 440206..440862 | + | 657 | ABQ56908 | 30S ribosomal protein S3 | - |
| LPC_3006 | 440879..441292 | + | 414 | ABQ56907 | 50S ribosomal protein L16/(L10E) | - |
| LPC_3005 | 441292..441486 | + | 195 | ABQ56906 | 50S ribosomal subunit protein L29 | - |
| LPC_3004 | 441488..441742 | + | 255 | ABQ56905 | 30S ribosomal protein S17 | - |
| LPC_3003 | 441831..442196 | + | 366 | ABQ56904 | 50S ribosomal protein L14 | - |
| LPC_3002 | 442209..442538 | + | 330 | ABQ56903 | 50S ribosomal protein L24 | - |
| LPC_3001 | 442554..443105 | + | 552 | ABQ56902 | 50S ribosomal protein L5 | - |
| LPC_3000 | 443118..443420 | + | 303 | ABQ56901 | 30S ribosomal protein S14 | - |
| LPC_2999 | 443449..443838 | + | 390 | ABQ56900 | 30S ribosomal protein S8 | - |
| LPC_2998 | 443856..444395 | + | 540 | ABQ56899 | 50S ribosomal protein L6/(L9E) | - |
| LPC_2997 | 444406..444765 | + | 360 | ABQ56898 | 50S ribosomal protein L18 | - |
| LPC_2996 | 444775..445281 | + | 507 | ABQ56897 | hypothetical protein | - |
| LPC_2995 | 445469..445903 | + | 435 | ABQ56896 | hypothetical protein | - |
| LPC_2994 | 445906..447234 | + | 1329 | ABQ56895 | preprotein translocase SecY | - |
| LPC_2993 | 447442..447798 | + | 357 | ABQ56894 | 30S ribosomal protein S13 | - |
| LPC_2992 | 447822..448220 | + | 399 | ABQ56893 | hypothetical protein | - |
| LPC_2991 | 448237..448857 | + | 621 | ABQ56892 | 30S ribosomal protein S4 | - |
| LPC_2990 | 448876..449868 | + | 993 | ABQ56891 | DNA-directed RNA polymerase alpha subunit RpoA | - |
| LPC_2989 | 449887..450270 | + | 384 | ABQ56890 | 50S ribosomal protein L17 | - |
| LPC_2988 | 450337..450816 | - | 480 | ABQ56889 | Single-strand binding protein (SSB) (Helix- destabilizing protein) | - |
| LPC_2987 | 450900..452267 | - | 1368 | ABQ56888 | major facilitator family transporter | - |
| LPC_2986 | 452429..453187 | + | 759 | ABQ56887 | glucose-1-dehydrogenase | - |
| LPC_2985 | 453258..453671 | + | 414 | ABQ56886 | acyl carrier protein | - |
| LPC_2984 | 453671..454567 | + | 897 | ABQ56885 | 3-hydroxymyristoyl/3-hydroxydecanoyl-(acyl carrier protein) dehydratase | - |
| LPC_2983 | 454572..455864 | + | 1293 | ABQ56884 | 3-oxoacyl-(acyl carrier protein) synthase II, C- terminal | - |
| LPC_2982 | 455865..457142 | + | 1278 | ABQ56883 | 3-oxoacyl-(acyl carrier protein) synthase II, N- terminal | - |
| LPC_2981 | 457135..457980 | + | 846 | ABQ56882 | lipid A biosynthesis acyltransferase | - |
| LPC_2980 | 458089..458394 | - | 306 | ABQ56881 | hypothetical protein | - |
| LPC_2979 | 458580..461270 | - | 2691 | ABQ56880 | hypothetical protein | - |
| LPC_2978 | 461583..462416 | - | 834 | ABQ56879 | diaminopimelate epimerase | - |
| LPC_2977 | 462713..463075 | - | 363 | ABQ56878 | hypothetical protein | - |
| LPC_2976 | 463265..463912 | + | 648 | ABQ56877 | carboxylesterase/phospholipase | - |
| LPC_2975 | 463909..464343 | + | 435 | ABQ56876 | oligoketide cyclase/lipid transporter protein | - |
| LPC_2974 | 464330..464602 | + | 273 | ABQ56875 | conserved hypothetical protein; UPF0125 | - |
| LPC_2973 | 464690..465034 | - | 345 | ABQ56874 | small protein A, tmRNA-binding | - |
| LPC_2972 | 465232..465642 | + | 411 | ABQ56873 | Ferric uptake regulation protein | - |
| LPC_2971 | 465780..465923 | + | 144 | ABQ56872 | hypothetical protein | - |
| LPC_2970 | 466082..467152 | + | 1071 | ABQ56871 | components of sensory transduction system | - |
| LPC_2969 | 467470..467856 | - | 387 | ABQ56870 | hypothetical protein | - |
| LPC_2968 | 468112..468711 | + | 600 | ABQ56869 | hypothetical protein | - |
| LPC_2967 | 468751..473040 | - | 4290 | ABQ56868 | SdhA, substrate of the Dot/Icm system | - |
| LPC_2966 | 473203..473922 | - | 720 | ABQ56867 | N-formylglutamate amidohydrolase | - |
| LPC_2965 | 473927..475147 | - | 1221 | ABQ56866 | conserved hypothetical protein; AF2307 from Archaeoglobus fulgidus | - |
| LPC_2964 | 475140..476594 | - | 1455 | ABQ56865 | ribosomal protein S6 modification protein | - |
| LPC_2963 | 476598..477311 | - | 714 | ABQ56864 | hypothetical protein conserved within Legionellae | - |
| LPC_2962 | 477506..477979 | - | 474 | ABQ56863 | conserved hypothetical protein | - |
| LPC_2961 | 477976..478527 | - | 552 | ABQ56862 | osmotically inducible protein Y | - |
| LPC_2960 | 478786..479244 | - | 459 | ABQ56861 | hypothetical protein conserved within Legionellae | - |
| LPC_2959 | 479339..482194 | + | 2856 | ABQ56860 | excinuclease ABC A subunit | - |
| LPC_2958 | 482378..482959 | + | 582 | ABQ56859 | LemA protein | - |
| LPC_2957 | 482970..483989 | + | 1020 | ABQ56858 | heat shock protein HtpX | - |
| LPC_2956 | 484016..484789 | - | 774 | ABQ56857 | ABC transporter, permease protein | - |
| LPC_2955 | 484779..485693 | - | 915 | ABQ56856 | ABC transporter, ATP binding component | - |
| LPC_2954 | 485924..486943 | - | 1020 | ABQ56855 | hypothetical protein | - |
| LPC_2953 | 487069..487707 | - | 639 | ABQ56854 | SM20-related protein | - |
| LPC_2952 | 487793..488500 | - | 708 | ABQ56853 | zinc metalloprotease | - |
| LPC_2951 | 488553..488666 | + | 114 | ABQ56852 | hypothetical protein | - |
| LPC_2950 | 488685..488966 | - | 282 | ABQ56851 | hypothetical protein | - |
| LPC_2949 | 489115..489978 | + | 864 | ABQ56850 | hypothetical protein | - |
| LPC_2948 | 490079..490546 | - | 468 | ABQ56849 | methylated DNA protein cysteine S- methyltransferase | - |
| LPC_2947 | 490550..490915 | - | 366 | ABQ56848 | 50S ribosomal protein L19 | - |
| LPC_2946 | 490942..491694 | - | 753 | ABQ56847 | tRNA (guanine N1) methyltransferase | - |
| LPC_2945 | 491694..492203 | - | 510 | ABQ56846 | 16S rRNA processing protein RimM | - |
| LPC_2944 | 492209..492469 | - | 261 | ABQ56845 | 30S ribosomal protein S16 | - |
| LPC_2943 | 492556..493932 | - | 1377 | ABQ56844 | signal recognition particle protein Ffh | - |
| LPC_2942 | 494190..494855 | - | 666 | ABQ56843 | hypothetical protein | - |
| LPC_2941 | 495128..496636 | + | 1509 | ABQ56842 | hypothetical protein | - |
| LPC_2940 | 496973..498376 | + | 1404 | ABQ56841 | Glutamate/gamma-aminobutyrate antiporter | - |
| LPC_2939 | 498432..498956 | + | 525 | ABQ56840 | hypothetical protein conserved within Legionellae | - |
| LPC_2938 | 499160..499501 | + | 342 | ABQ56839 | Carboxymuconolactone decarboxylase family | - |
| LPC_2937 | 499715..500158 | - | 444 | ABQ56838 | hypothetical protein conserved within Legionellae | - |
| LPC_2936 | 500177..500725 | - | 549 | ABQ56837 | inner (transmembrane) protein | - |
| LPC_2935 | 501142..501345 | - | 204 | ABQ56836 | hypothetical protein | - |
| LPC_2934 | 501413..502141 | + | 729 | ABQ56835 | conserved hypothetical protein | - |
| LPC_2933 | 502125..502670 | + | 546 | ABQ56834 | hypothetical protein conserved within Legionellae | - |
| LPC_2932 | 502675..503682 | + | 1008 | ABQ56833 | cytochrome c oxidase assembly protein | - |
| LPC_2931 | 503672..504556 | + | 885 | ABQ56832 | protoheme IX farnesyltransferase | - |
| LPC_2930 | 504566..505207 | + | 642 | ABQ56831 | hypothetical, SCO1/SenC family protein | - |
| LPC_2929 | 505573..506481 | + | 909 | ABQ56830 | glutathione synthase, ribosomal protein S6 modification protein | - |
| LPC_2928 | 506485..506730 | - | 246 | ABQ56829 | hypothetical protein conserved within Legionellae | - |
| LPC_2927 | 507048..508538 | + | 1491 | ABQ56828 | glucose-6-phosphate-1-dehydrogenase | - |
| LPC_2926 | 508525..509238 | + | 714 | ABQ56827 | 6-phosphogluconolactonase | - |
| LPC_2925 | 509226..511064 | + | 1839 | ABQ56826 | 6-phosphogluconate dehydratase | - |
| LPC_2924 | 511051..512046 | + | 996 | ABQ56825 | glucokinase | - |
| LPC_2923 | 512033..512695 | + | 663 | ABQ56824 | multifunctional: 2-keto-3-deoxygluconate 6- phosphate aldolase/(4-hydroxy-2-oxoglutarate aldolase | - |
| LPC_2922 | 512809..514230 | + | 1422 | ABQ56823 | D-xylose (galactose, arabinose)-proton symporter | - |
| LPC_2921 | 514320..515603 | - | 1284 | ABQ56822 | glucoamylase | - |
| LPC_2920 | 515779..516030 | - | 252 | ABQ56821 | transcriptional regulator, cro family | - |
| LPC_2919 | 516117..516656 | - | 540 | ABQ56820 | hypothetical protein | - |
| LPC_2918 | 516784..517779 | - | 996 | ABQ56819 | ferrochelatase | - |
| LPC_2917 | 517890..518123 | - | 234 | ABQ56818 | cold shock protein CspD | - |
| LPC_2916 | 518360..518791 | + | 432 | ABQ56817 | diadenosine tetraphosphate (Ap4A) hydrolase, histidine triad family protein | - |
| LPC_2915 | 519005..519433 | - | 429 | ABQ56816 | glyoxylase domain hypothetical protein | - |
| LPC_2914 | 519460..520803 | + | 1344 | ABQ56815 | outer membrane efflux protein | - |
| LPC_2913 | 520943..521980 | + | 1038 | ABQ56814 | multidrug resistance efflux pump PmrA | - |
| LPC_2912 | 521958..522890 | + | 933 | ABQ56813 | hypothetical protein | - |
| LPC_2911 | 522869..523756 | + | 888 | ABQ56812 | hypothetical protein | - |
| LPC_2910 | 523778..524158 | + | 381 | ABQ56811 | hypothetical protein | - |
| LPC_2909 | 524160..524591 | - | 432 | ABQ56810 | hypothetical protein | - |
| LPC_2908 | 524662..524751 | - | 90 | ABQ56809 | hypothetical protein | - |
| LPC_2907 | 524853..525464 | + | 612 | ABQ56808 | SAM-dependent methyltransferase | - |
| LPC_2906 | 525619..526419 | - | 801 | ABQ56807 | hypothetical protein | - |
| LPC_2905 | 526690..528690 | + | 2001 | ABQ56806 | hypothetical protein | - |
| LPC_2904 | 529509..530285 | - | 777 | ABQ56805 | hypothetical protein | - |
| LPC_2903 | 530651..530863 | + | 213 | ABQ56804 | hypothetical protein | - |
| LPC_2902 | 531033..531293 | + | 261 | ABQ56803 | IcmT | - |
| LPC_2901 | 531294..531638 | + | 345 | ABQ56802 | IcmS | - |
| LPC_2900 | 531738..532100 | + | 363 | ABQ56801 | IcmR | - |
| LPC_2899 | 532191..532766 | + | 576 | ABQ56800 | IcmQ | - |
| LPC_2898 | 532953..534083 | + | 1131 | ABQ56799 | IcmP (DotM) | - |
| LPC_2897 | 534080..536431 | + | 2352 | ABQ56798 | IcmO (DotL) | - |
| LPC_2896 | 536835..537404 | + | 570 | ABQ56797 | LphA (DotK) | - |
| LPC_2895 | 537416..537700 | + | 285 | ABQ56796 | IcmM (DotJ) | - |
| LPC_2894 | 537716..538354 | + | 639 | ABQ56795 | IcmL (DotI) | - |
| LPC_2893 | 538357..539439 | + | 1083 | ABQ56794 | IcmK (DotH) | - |
| LPC_2892 | 539444..542590 | + | 3147 | ABQ56793 | IcmE (DotG) | - |
| LPC_2891 | 542608..543414 | + | 807 | ABQ56792 | IcmG (DotF) | - |
| LPC_2890 | 543422..543763 | + | 342 | ABQ56791 | hypothetical protein | - |
| LPC_2889 | 544034..544432 | + | 399 | ABQ56790 | IcmD (DotP) | - |
| LPC_2888 | 544632..545258 | + | 627 | ABQ56789 | IcmJ (DotN) | - |
| LPC_2887 | 545294..548323 | + | 3030 | ABQ56788 | IcmB (DotO) | - |
| LPC_2886 | 548469..549725 | - | 1257 | ABQ56787 | proline/betaine transport protein like protein | - |
| LPC_2885 | 549728..552649 | - | 2922 | ABQ56786 | IcmF | - |
| LPC_2884 | 552649..553434 | - | 786 | ABQ56785 | IcmH (DotU) | - |
| LPC_2883 | 553719..555308 | - | 1590 | ABQ56784 | phosphoribosylamineimidazolecarboxamideformyltra nsferase | - |
| LPC_2882 | 555335..556204 | - | 870 | ABQ56783 | ribosomal protein L11 methyltransferase | - |
| LPC_2881 | 556206..557549 | - | 1344 | ABQ56782 | acetyl CoA carboxylase, biotin carboxylase subunit | - |
| LPC_2880 | 557562..558044 | - | 483 | ABQ56781 | acetyl-CoA carboxylase biotin carboxyl carrier protein | - |
| LPC_2879 | 558057..558494 | - | 438 | ABQ56780 | 3-dehydroquinate dehydratase type II | - |
| LPC_2878 | 558696..560486 | + | 1791 | ABQ56779 | oxaloacetate decarboxylase alpha subunit | - |
| LPC_2877 | 561052..562683 | + | 1632 | ABQ56778 | zinc metalloprotease | - |
| LPC_2876 | 562685..563533 | - | 849 | ABQ56777 | MhpC, Predicted hydrolases or acyltransferases (alpha/beta hydrolase superfamily) | - |
| LPC_2875 | 564002..564757 | + | 756 | ABQ56776 | endonuclease/exonuclease/phosphatase family protein | - |
| LPC_2874 | 564936..565946 | - | 1011 | ABQ56775 | fructose bisphosphate aldolase | - |
| LPC_2873 | 566096..566788 | - | 693 | ABQ56774 | phenol hydroxylase | - |
| LPC_2872 | 566914..567456 | + | 543 | ABQ56773 | IcmC (DotV)-like protein | - |
| LPC_2871 | 567658..567942 | - | 285 | ABQ56772 | conserved hypothetical protein | - |
| LPC_2870 | 568172..568909 | + | 738 | ABQ56771 | CDP-diacylglycerol-serine-O- phosphatidyltransferase | - |
| LPC_2869 | 569151..569420 | - | 270 | ABQ56770 | sugar transport PTS system phosphocarrier HPr protein | - |
| LPC_2868 | 569611..569910 | - | 300 | ABQ56769 | sigma-54 modulation protein | - |
| LPC_2867 | 569937..571331 | - | 1395 | ABQ56768 | RNA polymerase signma-54 factor RpoN | - |
| LPC_2866 | 571543..571707 | - | 165 | ABQ56767 | 50S ribosomal protein L33 | - |
| LPC_2865 | 571722..571958 | - | 237 | ABQ56766 | 50S ribosomal protein L28 | - |
| LPC_2864 | 572056..572217 | + | 162 | ABQ56765 | hypothetical protein | - |
| LPC_2863 | 572227..572904 | + | 678 | ABQ56764 | tRNA (guanine-N(7)-)-methyltransferase | - |
| LPC_2862 | 572916..574040 | + | 1125 | ABQ56763 | endo-1,4 beta-glucanase | - |
| LPC_2861 | 574118..575605 | + | 1488 | ABQ56762 | hypothetical protein | - |
| LPC_2860 | 575814..576956 | + | 1143 | ABQ56761 | protease subunit HflK | - |
| LPC_2859 | 576959..577873 | + | 915 | ABQ56760 | membrane protease subunit HflC | - |
| LPC_2858 | 578008..579303 | + | 1296 | ABQ56759 | Adenylosuccinate synthetase (IMP--aspartate ligase) (AdSS) (AMPSase) | - |
| LPC_2857 | 579812..580093 | + | 282 | ABQ56758 | hypothetical protein | - |
| LPC_2856 | 580293..580751 | - | 459 | ABQ56757 | arginine repressor | - |
| LPC_2855 | 580882..581616 | + | 735 | ABQ56756 | amino acid (glutamine) ABC transporter, periplasmic amino acid binding protein | - |
| LPC_2854 | 581613..582260 | + | 648 | ABQ56755 | amino acid (glutamine) ABC transporter, permease | - |
| LPC_2853 | 582244..582912 | + | 669 | ABQ56754 | amino acid (glutamine) ABC transporter, ATP binding component | - |
| LPC_2852 | 582909..584126 | + | 1218 | ABQ56753 | argininosuccinate synthase | - |
| LPC_2851 | 584119..585354 | + | 1236 | ABQ56752 | argininosuccinate lyase | - |
| LPC_2850 | 585357..586472 | + | 1116 | ABQ56751 | ornithine carbamoyltransferase | - |
| LPC_2849 | 586562..588037 | - | 1476 | ABQ56750 | adenosine deaminase | - |
| LPC_2848 | 588212..589327 | + | 1116 | ABQ56749 | leucine-, isoleucine-, valine-, threonine-, and alanine-binding protein | - |
| LPC_2847 | 589399..590736 | - | 1338 | ABQ56748 | carboxy-terminal protease | - |
| LPC_2846 | 590817..591959 | - | 1143 | ABQ56747 | Membrane-bound metallopeptidase | - |
| LPC_2845 | 591946..593490 | - | 1545 | ABQ56746 | phosphoglycerate mutase, 2,3-bisphosphoglycerate independent | - |
| LPC_2844 | 593593..593889 | - | 297 | ABQ56745 | hypothetical protein | - |
| LPC_2843 | 593966..595033 | - | 1068 | ABQ56744 | hypothetical protein | - |
| LPC_2842 | 595584..596300 | + | 717 | ABQ56743 | undecaprenyl diphosphate synthetase | - |
| LPC_2841 | 596448..597110 | + | 663 | ABQ56742 | phosphatidate cytidyltransferase | - |
| LPC_2840 | 597143..598495 | + | 1353 | ABQ56741 | membrane associated zinc metalloprotease | - |
| LPC_2839 | 598666..600978 | + | 2313 | ABQ56740 | outer membrane protein | - |
| LPC_2838 | 601092..601592 | + | 501 | ABQ56739 | outer membrane protein OmpH | - |
| LPC_2837 | 601594..602625 | + | 1032 | ABQ56738 | UDP-3-O-(3-hydroxymyristoyl) glucosamine N- acyltransferase | - |
| LPC_2836 | 602743..603195 | + | 453 | ABQ56737 | hypothetical protein | - |
| LPC_2835 | 603192..603962 | + | 771 | ABQ56736 | acyl-(acyl carrier protein)-UDP-N- acetylglucosamine acyltransferase | - |
| LPC_2834 | 603966..604370 | + | 405 | ABQ56735 | CrcB protein, camphor resistance | - |
| LPC_2833 | 604392..605672 | + | 1281 | ABQ56734 | seryl tRNA synthetase | - |
| LPC_2832 | 605811..606035 | + | 225 | ABQ56733 | hypothetical protein | - |
| LPC_2831 | 606206..606469 | + | 264 | ABQ56732 | hypothetical protein | - |
| LPC_2830 | 606664..606828 | + | 165 | ABQ56731 | hypothetical protein | - |
| LPC_2829 | 606809..607741 | + | 933 | ABQ56730 | hypothetical protein | - |
| LPC_2828 | 607777..608589 | + | 813 | ABQ56729 | hypothetical protein | - |
| LPC_2827 | 608579..609409 | - | 831 | ABQ56728 | aldo/keto reductase-like diketogulonate reductase | - |
| LPC_2826 | 609848..610696 | - | 849 | ABQ56727 | hypothetical protein | - |
| LPC_2825 | 611033..611500 | - | 468 | ABQ56726 | hypothetical protein | - |
| LPC_2824 | 611484..612242 | - | 759 | ABQ56725 | methyltransferase, ubiE/COQ5 family | - |
| LPC_2823 | 612208..612816 | - | 609 | ABQ56724 | hypothetical protein | - |
| LPC_2822 | 612813..613280 | - | 468 | ABQ56723 | acetyltransferase, GNAT family | - |
| LPC_2821 | 613277..613435 | - | 159 | ABQ56722 | hypothetical protein | - |
| LPC_2820 | 613432..613995 | - | 564 | ABQ56721 | aminoglycoside N (6')-acetyltransferase | - |
| LPC_2819 | 614497..615231 | + | 735 | ABQ56720 | hypothetical protein | - |
| LPC_2818 | 615234..616457 | + | 1224 | ABQ56719 | site specific recombinase | - |
| LPC_2817 | 616462..616881 | + | 420 | ABQ56718 | hypothetical protein | - |
| LPC_2816 | 616983..617657 | - | 675 | ABQ56717 | hypothetical protein | - |
| LPC_2815 | 617842..618723 | + | 882 | ABQ56716 | Legionella vir region protein | - |
| LPC_2814 | 618740..619048 | + | 309 | ABQ56715 | Legionella vir region protein | - |
| LPC_2813 | 619069..619335 | + | 267 | ABQ56714 | global regulator (carbon storage regulator) | - |
| LPC_2812 | 619346..620311 | + | 966 | ABQ56713 | LvhB11 | - |
| LPC_2811 | 620323..620700 | + | 378 | ABQ56712 | hypothetical protein | - |
| LPC_2810 | 620751..620996 | + | 246 | ABQ56711 | Conjugal transfer protein trbD | - |
| LPC_2809 | 620993..623518 | + | 2526 | ABQ56710 | Conjugal transfer protein trbE precursor | - |
| LPC_2808 | 623515..624258 | + | 744 | ABQ56709 | Conjugal transfer protein trbF | - |
| LPC_2807 | 624255..625130 | + | 876 | ABQ56708 | Conjugal transfer protein trbG precursor | - |
| LPC_2806 | 625133..625543 | + | 411 | ABQ56707 | hypothetical protein | - |
| LPC_2805 | 625549..626799 | + | 1251 | ABQ56706 | Conjugal transfer protein trbI | - |
| LPC_2804 | 626815..627555 | + | 741 | ABQ56705 | Conjugal transfer protein trbJ precursor | - |
| LPC_2803 | 627582..627794 | + | 213 | ABQ56704 | hypothetical protein | - |
| LPC_2802 | 627799..629196 | + | 1398 | ABQ56703 | Conjugal transfer protein trbL | - |
| LPC_2801 | 629193..631082 | + | 1890 | ABQ56702 | Conjugal transfer protein traG | - |
| LPC_2800 | 631079..631624 | + | 546 | ABQ56701 | Plasmid transfer protein traF precursor | - |
| LPC_2799 | 631621..631785 | + | 165 | ABQ56700 | traD protein | - |
| LPC_2798 | 631778..633964 | + | 2187 | ABQ56699 | DNA primase traC (Replication primase) | - |
| LPC_2797 | 633966..634424 | - | 459 | ABQ56698 | hypothetical protein | - |
| LPC_2796 | 634435..635202 | - | 768 | ABQ56697 | hypothetical protein | - |
| LPC_2795 | 635310..637175 | - | 1866 | ABQ56696 | traI protein | - |
| LPC_2794 | 637172..637525 | - | 354 | ABQ56695 | traJ protein | - |
| LPC_2793 | 637940..638266 | + | 327 | ABQ56694 | TraK | - |
| LPC_2792 | 638266..638991 | + | 726 | ABQ56693 | traL protein | - |
| LPC_2791 | 638991..639440 | + | 450 | ABQ56692 | traM protein | - |
| LPC_2790 | 639466..641496 | + | 2031 | ABQ56691 | Putative type I restriction enzyme HindVIIP M protein | - |
| LPC_2789 | 641493..642767 | + | 1275 | ABQ56690 | hypothetical protein | - |
| LPC_2788 | 642764..645745 | + | 2982 | ABQ56689 | Putative type I restriction enzyme R protein | - |
| LPC_2787 | 645773..647026 | + | 1254 | ABQ56688 | hypothetical protein | - |
| LPC_2786 | 647023..648783 | + | 1761 | ABQ56687 | hypothetical protein | virb4 |
| LPC_2785 | 648787..652206 | + | 3420 | ABQ56686 | putative RNA helicase | - |
| LPC_2784 | 652210..652533 | - | 324 | ABQ56685 | hypothetical protein | - |
Host bacterium
| ID | 99 | Element type | ICE (Integrative and conjugative element) |
| Element name | Trb-1 | GenBank | CP000675 |
| Element size | 3576470 bp | Coordinate of oriT [Strand] | 637526..637939 [+] |
| Host bacterium | Legionella pneumophila str. Corby | Coordinate of element | 614497..656813 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |