Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200090 |
| Name | oriT_ICEclc-1 |
| Organism | Pseudomonas putida clc |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | AJ617740 (51849..52332 [+], 484 nt) |
| oriT length | 484 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | (1) In oriT_ICEclc, there is a dual oriT system. In this system, both oriTs are functional and compared to the ICEclc only harbors one oriT, there is a fourfold increase of transfer rate in ICEclc that habors the dual oriT system [PMID:21255116, PMID:22016851]. (2) The relaxase orf50240_ICEclc processes both oriTs in a different manner. For nicking-rejoining oriT_ICEclc-2, other factors would be required [PMID:21255116, PMID:22016851]. |
oriT sequence
Download Length: 484 nt
>oriT_ICEclc-1
GAGGCGGGAAACCGCTGGACCAGCTCGGCGTAGCGCTCCAGCGGTGCGCGATAGAGGGTGGCGAACTGCTTGCGCGACAGCGACGTGCGCTGCCAGATGTGTTCCAGTAGCTTCTGCCGGCGCGGTGTCGCCAGCAGCGAGGCGGCCGACTCGGGCCGCAGCCGCCCTTTCGGGAGGTCGTTGGAGGGCGCTGGCGACGGAGCAGAAGCGACCACGGGCCGTTTTCGCTGGAACAGAGAGAGCATGTGGGTGTCCTAGTGGCGGGCCGACCGGGAGGCCTTTTGCCTTTTCGAGGTAGGGCCTTTCCCCTTGCAGCCCCTTCCATTGCCCTTTCGGCCCTTTGGCCTTTAACCATTTGGATATAGGGCGGCGTGCGCTTGCCGTCCACCACCAATGCGGGTTGCAGCGAGCCGGATTGGTGCGTGAAGGCTCGCTGTTCCCCCCCATACGATGGCCCGATTCTTGGACTCTGGAGAGCATCATG
GAGGCGGGAAACCGCTGGACCAGCTCGGCGTAGCGCTCCAGCGGTGCGCGATAGAGGGTGGCGAACTGCTTGCGCGACAGCGACGTGCGCTGCCAGATGTGTTCCAGTAGCTTCTGCCGGCGCGGTGTCGCCAGCAGCGAGGCGGCCGACTCGGGCCGCAGCCGCCCTTTCGGGAGGTCGTTGGAGGGCGCTGGCGACGGAGCAGAAGCGACCACGGGCCGTTTTCGCTGGAACAGAGAGAGCATGTGGGTGTCCTAGTGGCGGGCCGACCGGGAGGCCTTTTGCCTTTTCGAGGTAGGGCCTTTCCCCTTGCAGCCCCTTCCATTGCCCTTTCGGCCCTTTGGCCTTTAACCATTTGGATATAGGGCGGCGTGCGCTTGCCGTCCACCACCAATGCGGGTTGCAGCGAGCCGGATTGGTGCGTGAAGGCTCGCTGTTCCCCCCCATACGATGGCCCGATTCTTGGACTCTGGAGAGCATCATG
oriT secondary structure
Predicted by RNAfold.
Download structure file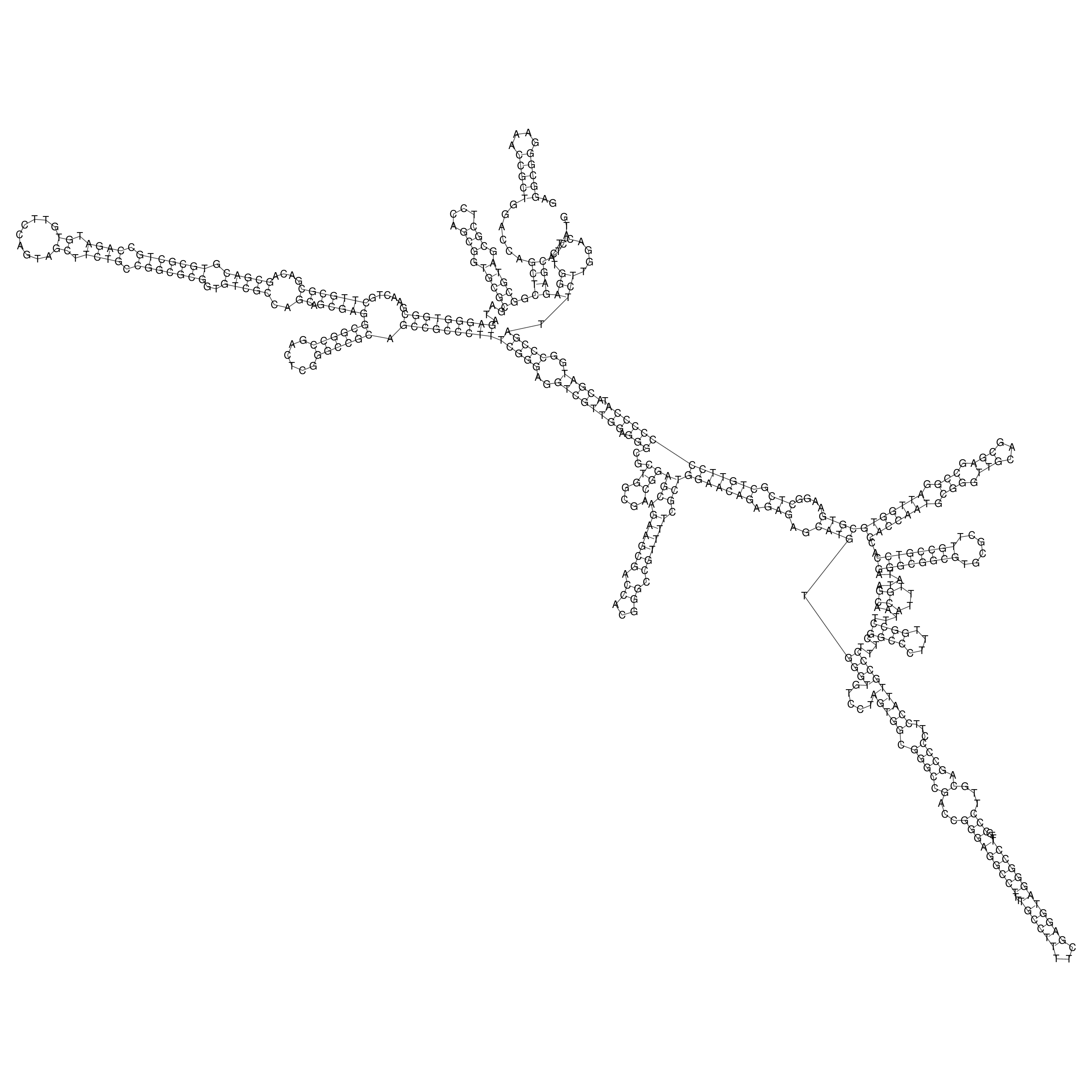
Reference
[1] Ryo Miyazaki et al. (2011) A dual functional origin of transfer in the ICEclc genomic island of Pseudomonas knackmussii B13. Molecular microbiology. 79(3):743-58. [PMID:21255116]
Relaxase
| ID | 1094 | GenBank | CAE92900 |
| Name | orf50240_ICEclc |
UniProt ID | Q706Q0 |
| Length | 615 a.a. | PDB ID | |
| Note | |||
Relaxase protein sequence
Download Length: 615 a.a. Molecular weight: 67419.64 Da Isoelectric Point: 7.4238
>CAE92900.1 hypothetical protein [Pseudomonas putida]
MLSLFQRKRPVVASAPSPAPSNDLPKGRLRPESAASLLATPRRQKLLEHIWQRTSLSRKQFATLYRAPLE
RYAELVQRFPASEAHHHAYPGGMLDHGLEIVAYALKLRQSHLLPAGSTPEDQAAQSEAWTAAVAYAALLH
DVGKLAVDLHVELADGSTWHPWHGPLRQPYRFRYREDREYRLHSAATGLLYRQLLDAQLLDWLSGYPALW
GPLLYVLAGQYEHAGVLGELVVQADRASVAQELGGDPARAMAAPKHALQRKLLDGLRYLLKEELKLNQPE
ASDGWLTEDALWLVSKTVSDKLRAHLLSQGIDGIPANNTAVFNVLQDHGMLHPTTDGKAVWRATVTSATG
WSHSFTLLRLAPALIWEPGERPVPFAGTVAIDAVPNDKSVAQPAAVAETTQEGQEAPPWESSSVAVPSPA
TQTVPDVLEDMLAMVGMGNSSGTQQDEEATSHAADATPSEAPMPAMAAASPPSPVPPAAPSSATAQPSGE
HFMAWLKQGIVSRRLIINDAKALVHTVSDTAYLVSPGVFQRYAREHPQVGMLAKQESQQDWQWVQKRFEK
LQLHRKHINGLNVWACSVTGSRKSRQLHGYLLRDPLPLFGKVPPNNPYLSILQSI
MLSLFQRKRPVVASAPSPAPSNDLPKGRLRPESAASLLATPRRQKLLEHIWQRTSLSRKQFATLYRAPLE
RYAELVQRFPASEAHHHAYPGGMLDHGLEIVAYALKLRQSHLLPAGSTPEDQAAQSEAWTAAVAYAALLH
DVGKLAVDLHVELADGSTWHPWHGPLRQPYRFRYREDREYRLHSAATGLLYRQLLDAQLLDWLSGYPALW
GPLLYVLAGQYEHAGVLGELVVQADRASVAQELGGDPARAMAAPKHALQRKLLDGLRYLLKEELKLNQPE
ASDGWLTEDALWLVSKTVSDKLRAHLLSQGIDGIPANNTAVFNVLQDHGMLHPTTDGKAVWRATVTSATG
WSHSFTLLRLAPALIWEPGERPVPFAGTVAIDAVPNDKSVAQPAAVAETTQEGQEAPPWESSSVAVPSPA
TQTVPDVLEDMLAMVGMGNSSGTQQDEEATSHAADATPSEAPMPAMAAASPPSPVPPAAPSSATAQPSGE
HFMAWLKQGIVSRRLIINDAKALVHTVSDTAYLVSPGVFQRYAREHPQVGMLAKQESQQDWQWVQKRFEK
LQLHRKHINGLNVWACSVTGSRKSRQLHGYLLRDPLPLFGKVPPNNPYLSILQSI
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q706Q0 |
Reference
[1] Ryo Miyazaki et al. (2011) A dual functional origin of transfer in the ICEclc genomic island of Pseudomonas knackmussii B13. Molecular microbiology. 79(3):743-58. [PMID:21255116]
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 53593..74302
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_48 | 48928..49731 | + | 804 | CAE92899 | putative transcriptional regulator | - |
| Locus_49 | 50246..52093 | - | 1848 | CAE92900 | hypothetical protein | - |
| Locus_50 | 52330..52716 | + | 387 | CAE92901 | hypothetical protein | - |
| Locus_51 | 52716..53174 | + | 459 | CAE92902 | hypothetical protein | - |
| Locus_52 | 53202..53579 | + | 378 | CAE92903 | hypothetical protein | - |
| Locus_53 | 53593..55110 | - | 1518 | CAE92904 | hypothetical protein | tfc19 |
| Locus_54 | 55126..55485 | - | 360 | CAE92905 | hypothetical protein | tfc18 |
| Locus_55 | 55482..56879 | - | 1398 | CAE92906 | hypothetical protein | tfc22 |
| Locus_56 | 56889..57836 | - | 948 | CAE92907 | hypothetical protein | tfc23 |
| Locus_57 | 57833..58279 | - | 447 | CAE92908 | hypothetical protein | tfc24 |
| Locus_58 | 58438..58932 | - | 495 | CAE92909 | putative DNA repair protein | - |
| Locus_59 | 59116..59880 | - | 765 | CAE92910 | putative protein-disulfide isomerase | - |
| Locus_60 | 59894..62761 | - | 2868 | CAE92911 | conserved hypothetical protein with VirB4 domain | virb4 |
| Locus_61 | 62761..63201 | - | 441 | CAE92912 | hypothetical protein | tfc15 |
| Locus_62 | 63182..64600 | - | 1419 | CAE92913 | hypothetical protein | tfc14 |
| Locus_63 | 64590..65522 | - | 933 | CAE92914 | hypothetical protein | tfc13 |
| Locus_64 | 65519..66211 | - | 693 | CAE92915 | hypothetical protein | tfc12 |
| Locus_65 | 66208..66618 | - | 411 | CAE92916 | hypothetical protein | tfc11 |
| Locus_66 | 66631..66990 | - | 360 | CAE92917 | hypothetical protein | tfc10 |
| Locus_67 | 67007..67240 | - | 234 | CAE92918 | hypothetical protein | tfc9 |
| Locus_68 | 67237..67620 | - | 384 | CAE92919 | hypothetical protein | tfc8 |
| Locus_69 | 67806..68210 | + | 405 | CAE92920 | hypothetical protein | - |
| Locus_70 | 68247..68996 | - | 750 | CAE92921 | hypothetical protein | tfc7 |
| Locus_71 | 68993..71179 | - | 2187 | CAE92922 | hypothetical protein | - |
| Locus_72 | 71184..71732 | - | 549 | CAE92923 | hypothetical protein | tfc5 |
| Locus_73 | 71729..72319 | - | 591 | CAE92924 | hypothetical protein | virB1 |
| Locus_74 | 72301..73020 | - | 720 | CAE92925 | hypothetical protein | tfc3 |
| Locus_75 | 73035..73685 | - | 651 | CAE92926 | hypothetical protein | - |
| Locus_76 | 73682..74302 | - | 621 | CAE92927 | hypothetical protein | tfc2 |
| Locus_77 | 74442..75311 | - | 870 | CAE92928 | hypothetical protein | - |
| Locus_78 | 75425..77704 | - | 2280 | CAE92929 | putative DNA/RNA helicase | - |
| Locus_79 | 77804..78913 | - | 1110 | CAE92930 | hypothetical protein | - |
Host bacterium
| ID | 98 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEclc | GenBank | AJ617740 |
| Element size | 105032 bp | Coordinate of oriT [Strand] | 51849..52332 [+]; 94918..95237 [+] |
| Host bacterium | Pseudomonas putida clc |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | bphA1 |
| Symbiosis gene | - |
| Anti-CRISPR | - |