Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200081 |
| Name | oriT_ICEMlSym |
| Organism | Mesorhizobium loti R7A |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | AL672111 (133568..133628 [+], 61 nt) |
| oriT length | 61 nt |
| IRs (inverted repeats) | 13..31, 36..54 (AGACAAAGGCTTCGCCGCC..GGAGGCGGTGCCTTTATCT) |
| Location of nic site | 58..59 |
| Conserved sequence flanking the nic site |
ATCTTGCC|T |
| Note |
oriT sequence
Download Length: 61 nt
>oriT_ICEMlSym
AAAACGATGTGCAGACAAAGGCTTCGCCGCCTCATGGAGGCGGTGCCTTTATCTTGCCTTA
AAAACGATGTGCAGACAAAGGCTTCGCCGCCTCATGGAGGCGGTGCCTTTATCTTGCCTTA
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file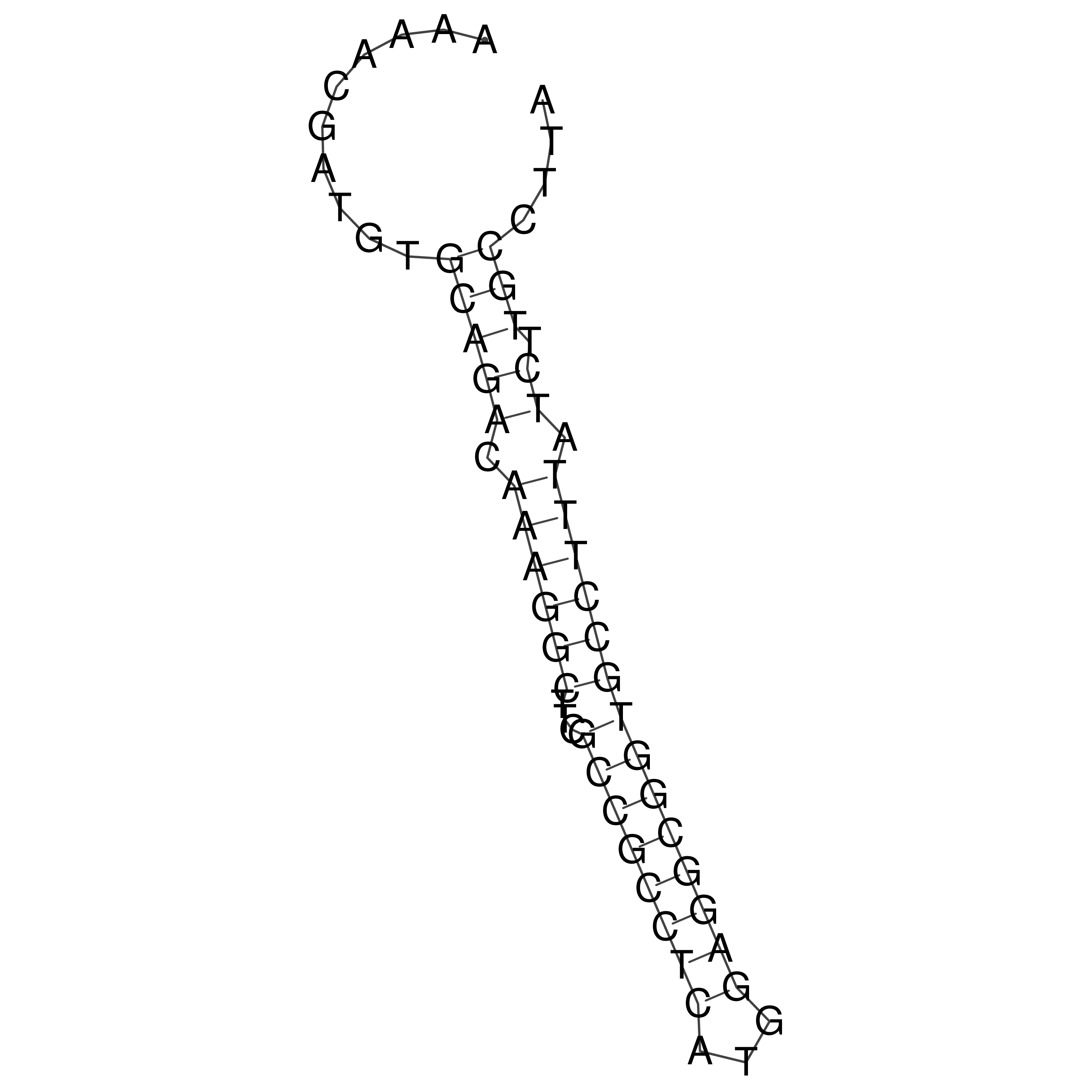
Reference
[1] Callum J Verdonk et al. (2019) Delineation of the integrase-attachment and origin-of-transfer regions of the symbiosis island ICEMlSymR7A. Plasmid. 104:102416. [PMID:31078551]
Relaxase
| ID | 1070 | GenBank | WP_029354922 |
| Name | RlxS_ICEMlSym |
UniProt ID | A0A3S3MR69 |
| Length | 657 a.a. | PDB ID | |
| Note | |||
Relaxase protein sequence
Download Length: 657 a.a. Molecular weight: 73054.27 Da Isoelectric Point: 9.9445
>WP_029354922.1 MULTISPECIES: relaxase/mobilization nuclease RlxS [Mesorhizobium]
MADDEFRPKLGKIGSRGSKAGKRYAGQVRAAVNRAGGQPQPGGRFTGSRTGRGGAAAALLKSRDRYAAFR
QRRVIVKARIVKLAGKGADGARAHLRYLQRDGVTREGAPGELYGADSDSVDGKAFIDRADGDRHQFRFIV
AAEDGIEYEDLKPLTRRLMAQMQEDLGTKLDWVAVDHFNTGHPHSHIIVRGRDGRGENLVIAREYISSGI
RERAAELVSLDLGPRTDREIELRLRREMEQERFTSIDRRLLSMRDDDGLVSPDGPDAIRQTLHQGRLRKL
ERMGLAAEVGAGSWRLDDELEATLRRTGERGDIIKTMHRELAGKGVARSAADWVIHDRAGEPVQSLVGRV
VARGLADEINDRHYMIVDAVDGKSHWIDIGRGEAMETMPNGCIVRLAPRNTEPRQVDRNIAEIAAAHGGR
YDVDIHLKHDPSATESFARTHVRRLEAIRRATGGVEREPNGTWLIAPDHLDRVANYEGQRARAEPVVVDK
LSSMALERQVSFDGATWLDRELVADRPEPLHGSGFGRDVREAQARRRQWLIAQGLAYEEQDRIVYRANML
SILRQRELNRVAGQLSEELGLPYAEARSGGRVEGTLRRSVELASGKYAVVEKSREFTLVPWRPVLERHVG
KEVSGVVSGEGEISWTVGRQRSGPGVS
MADDEFRPKLGKIGSRGSKAGKRYAGQVRAAVNRAGGQPQPGGRFTGSRTGRGGAAAALLKSRDRYAAFR
QRRVIVKARIVKLAGKGADGARAHLRYLQRDGVTREGAPGELYGADSDSVDGKAFIDRADGDRHQFRFIV
AAEDGIEYEDLKPLTRRLMAQMQEDLGTKLDWVAVDHFNTGHPHSHIIVRGRDGRGENLVIAREYISSGI
RERAAELVSLDLGPRTDREIELRLRREMEQERFTSIDRRLLSMRDDDGLVSPDGPDAIRQTLHQGRLRKL
ERMGLAAEVGAGSWRLDDELEATLRRTGERGDIIKTMHRELAGKGVARSAADWVIHDRAGEPVQSLVGRV
VARGLADEINDRHYMIVDAVDGKSHWIDIGRGEAMETMPNGCIVRLAPRNTEPRQVDRNIAEIAAAHGGR
YDVDIHLKHDPSATESFARTHVRRLEAIRRATGGVEREPNGTWLIAPDHLDRVANYEGQRARAEPVVVDK
LSSMALERQVSFDGATWLDRELVADRPEPLHGSGFGRDVREAQARRRQWLIAQGLAYEEQDRIVYRANML
SILRQRELNRVAGQLSEELGLPYAEARSGGRVEGTLRRSVELASGKYAVVEKSREFTLVPWRPVLERHVG
KEVSGVVSGEGEISWTVGRQRSGPGVS
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3S3MR69 |
Reference
[1] Callum J Verdonk et al. (2019) Delineation of the integrase-attachment and origin-of-transfer regions of the symbiosis island ICEMlSymR7A. Plasmid. 104:102416. [PMID:31078551]
Host bacterium
| ID | 89 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEMlSym | GenBank | AL672111 |
| Element size | 501801 bp | Coordinate of oriT [Strand] | 133568..133628 [+] |
| Host bacterium | Mesorhizobium loti R7A |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | btpB, gmd |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixS, fixI, fixH, fixG, fixP, fixQ, fixO, fixN, nodM, nodD, nolL, nodB, eB234_29755, nodJ, nodI, nodC, nodA, nodS, mlr6097, nifU, nifX, nifN, nifE, nifK, nifD, nifH, nifQ, mLTONO_5203, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, nifT, nolK, noeL, nodZ |
| Anti-CRISPR | AcrIIC1, AcrIIA7 |