Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200080 |
| Name | oriT_GGI |
| Organism | Neisseria gonorrhoeae strain MS11 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | AY803022 (6050..6099 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 13..25, 32..44 (CAAAGGCCTGCAT..ATGCAGGCCTTTG) |
| Location of nic site | 46..47 |
| Conserved sequence flanking the nic site |
TTGCCT|A |
| Note | The MOBH family relaxase TraI_GGI cleaves oriT_GGI in a site-specific and Mn2+-dependent manner [PMID:21255116, PMID:22016851]. |
oriT sequence
Download Length: 50 nt
TTAAAGGTAAAACAAAGGCCTGCATGTAAAAATGCAGGCCTTTGCCTATT
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file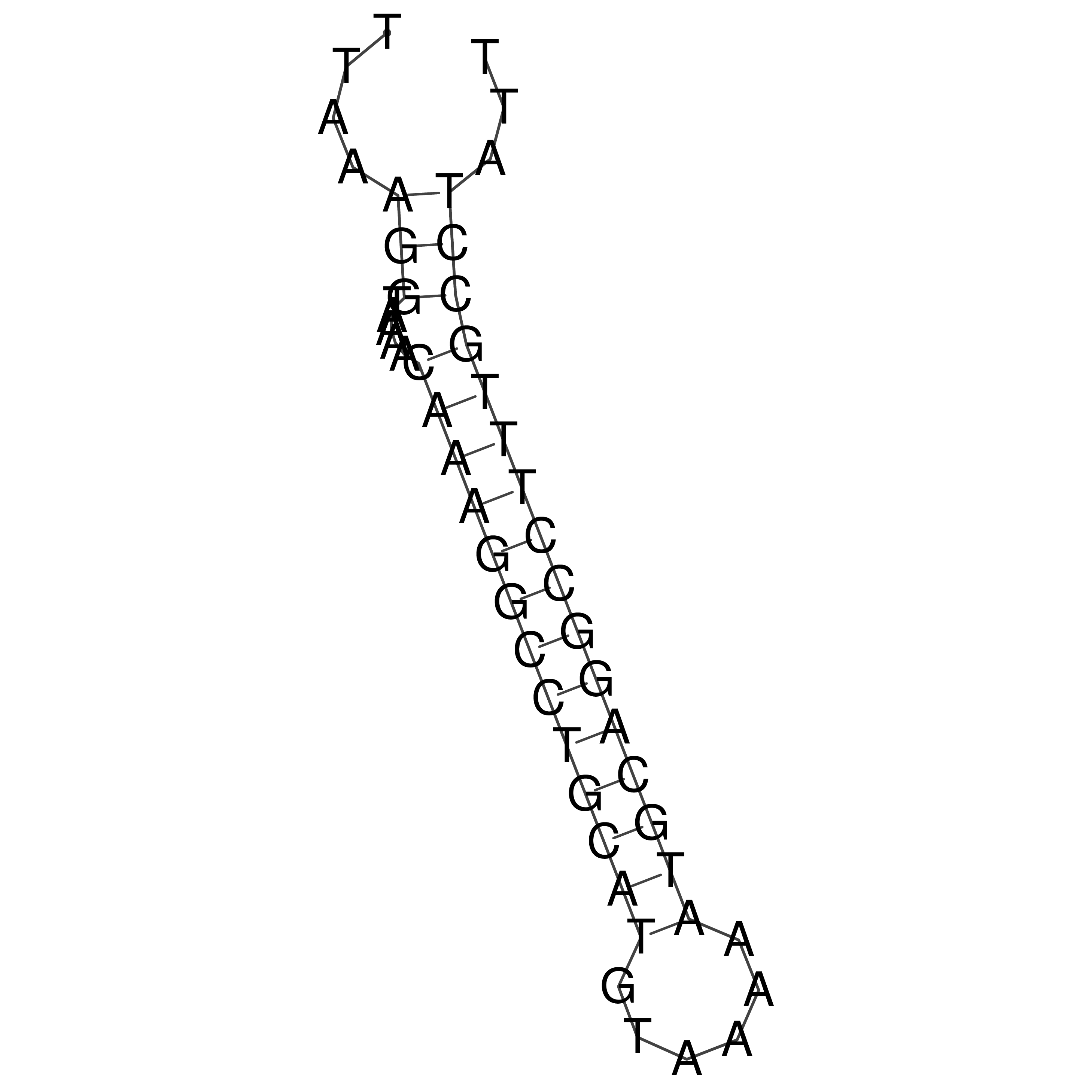
Reference
[1] Jan-Hendrik Heilers et al. (2019) DNA processing by the MOBH family relaxase TraI encoded within the gonococcal genetic island. Nucleic acids research. 47(15):8136-8153. [PMID:31276596]
Relaxase
| ID | 1069 | GenBank | WP_003693100 |
| Name | TraI_GGI |
UniProt ID | D6H7W1 |
| Length | 850 a.a. | PDB ID | |
| Note | The MOBH family relaxase TraI_GGI cleaves oriT_GGI in a site-specific and Mn2+-dependent manner [PMID:21255116, PMID:22016851]. | ||
Relaxase protein sequence
Download Length: 850 a.a. Molecular weight: 94671.82 Da Isoelectric Point: 5.9293
MKTSLLTIASSLMLVSGSMLYLINTRKSTDSVSHNDEVAHINQNIVNDRFRILNAHELIQVLDLSPQISG
IKMNLGLSDENWSKDALPFLEKYIAFVQRLPASESHHHAGDGGLVRHTLDVAALALVASTSQSWPPNAKT
EEIAKKTAVWRYGIMCAALLHDVGKTVTGFQVELFDSAISLEKLLWLPDTGSMAESGKLYYRVEFPDAKS
AYSTHAEIAWTFFQALVPSHVRQWLATTDPNLMITLRNYLSGKKDGSPLEQLIKNADMTSVSRDLRSGSR
QRFSTAKRKPFIETIMETLKEMLSDRGVHFSIATTAGGDLFRKGDRIYLMSKNVPDYIRQYLRKNQHPAA
GSFPADNQRIFDTLFEYRAVIPPENDPHRAINHIEVEFTRMDGEKVRNIFSVLCFNAKTLYPDGDYPTEF
LGNLSEVSAKKEIPDVVVSEDKVSTQIQGAEDNPVTMTKDKMDFLSIQPVTAEQPPLPEKNQTETANSAF
DNSMNTAAPAENKSSESEKAKEPGYGTIDNLIENFNLIEADSEQTESTKSEAETAENAVNLVQENIRVEV
KTETVDKPRAEVKETETDKAPKTIKSGTKASRKKLAGLFADNRKPIGKTAEPDAQIKTTETVTHQIDSEA
VSLPESESAQNGMRELERLRLREPTSSPRPVQVMDDSEGLDSLASGVKNMAELEAIVSERNLKKQSETES
TEIVEDSVKAKHMEARDRGMQFLRWLADGLGDGSIAVNRSGATVHFIEQGMLLVTPAVFRDYAGGVFNKS
DTESLGPLTQKGFEALRLHARTKKSSLHRVLTTNGSNRRLFYCYLIPEHNIKHIIQPGSRPQNNTDIKLD
ESDLLVMEQK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | D6H7W1 |
Reference
[1] Jan-Hendrik Heilers et al. (2019) DNA processing by the MOBH family relaxase TraI encoded within the gonococcal genetic island. Nucleic acids research. 47(15):8136-8153. [PMID:31276596]
T4CP
| ID | 647 | GenBank | AAW83117 |
| Name | ParA_GGI |
UniProt ID | Q5EP68 |
| Length | 295 a.a. | PDB ID | _ |
| Note | |||
T4CP protein sequence
Download Length: 295 a.a. Molecular weight: 32561.85 Da Isoelectric Point: 8.0343
MSAPVILSAAATKGGVGKTTLIANVSAVLADIGLRVLMIDCDVQPSLSKYYPISHRAPNGIVELLLGENT
EEIIRSTISNTVFPNLDIVISNNISAFVETEVTSRPDRAFLLKSKLHSQYIQNNYDVVLIDTQGAVGPLL
DTAAFAATCLLCPIMPEVLSAREFLTGTQELLKRLQKGEAMRIPMPQLRAIIYAQDRTNDAKFIASSIKD
FFSNTLDDRRKLLSVAVPAAKAYKEATTLRVPVHCHERKQLGKMLPAYEIMHRIVYEIFPGIEQKNLRGS
CFNDMASLLQKEVQS
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q5EP68 |
Reference
[1] Melanie M Callaghan et al. (2021) Expression, Localization, and Protein Interactions of the Partitioning Proteins in the Gonococcal Type IV Secretion System. Frontiers in microbiology. 12:784483. [PMID:34975804]
| ID | 648 | GenBank | AAW83116 |
| Name | ParB_GGI |
UniProt ID | Q5EP69 |
| Length | 496 a.a. | PDB ID | _ |
| Note | |||
T4CP protein sequence
Download Length: 496 a.a. Molecular weight: 56519.41 Da Isoelectric Point: 4.7899
MNLDQNKATLSLHIGTPAQTNMSFGQEDTPSRGMLMSLPISEIDFFDKNPRRLHDVSSYAAIKESIRSSG
IQQPVHVTQRPGKSKYVLAQGGNTRLKILCELLEETGDNRFRMMPCIYIPYSDETTLQVAHLVENELRAE
MCFWDKACKYAEIRDIFQDESDSGKLSLRELEVMFSDKGLTVSHKTLGLFFFASDNLQELQEYAFKLSHA
KVEEIKKLHSTLFSISKSIQLDDSFQYLWSDVLEQWHNQYSATELDVHELYQFVLDRFKFEYGSDAFSET
ENSSDKDKIETAAKQDVNLHQKDSDMEFVPAKVNGTKADAEDEPKEAEPITLEQTSKKLFSAVRKLLAKV
SLSDCFKIHKDFPYGWYVEYPAFEHISASSAGNAIYAIDVLHEEAGNVFTFLWKLSGIDTFMFDVGDSSN
PLLTLKSDSKLRIAYEDPDKYDEYTNCGIGNKENIIASLLNWQTNTSHEFHANIVEILSLSTQYNNLRKT
GAKSEE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q5EP69 |
Reference
[1] Melanie M Callaghan et al. (2021) Expression, Localization, and Protein Interactions of the Partitioning Proteins in the Gonococcal Type IV Secretion System. Frontiers in microbiology. 12:784483. [PMID:34975804]
Host bacterium
| ID | 88 | Element type | ICE (Integrative and conjugative element) |
| Element name | GGI | GenBank | AY803022 |
| Element size | 57358 bp | Coordinate of oriT [Strand] | 6050..6099 [-] |
| Host bacterium | Neisseria gonorrhoeae strain MS11 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIB |