Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200052 |
| Name | oriT_Tn6079 |
| Organism | Uncultured bacterium MID12 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | GU951538 (7612..7763 [+], 152 nt) |
| oriT length | 152 nt |
| IRs (inverted repeats) | 3..7, 19..23 (CCCCT..AGGGG) 60..71, 76..87 ( GAAAATCCTTTG..CAAGGGATTTAC) |
| Location of nic site | 72..73nt |
| Conserved sequence flanking the nic site |
TGG|T |
| Note | blastn alignment with the oriT_Tn916 |
oriT sequence
Download Length: 152 nt
>oriT_Tn6079
AACCCCTCGGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGATTGGAGGGATTT
AACCCCTCGGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGATTGGAGGGATTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file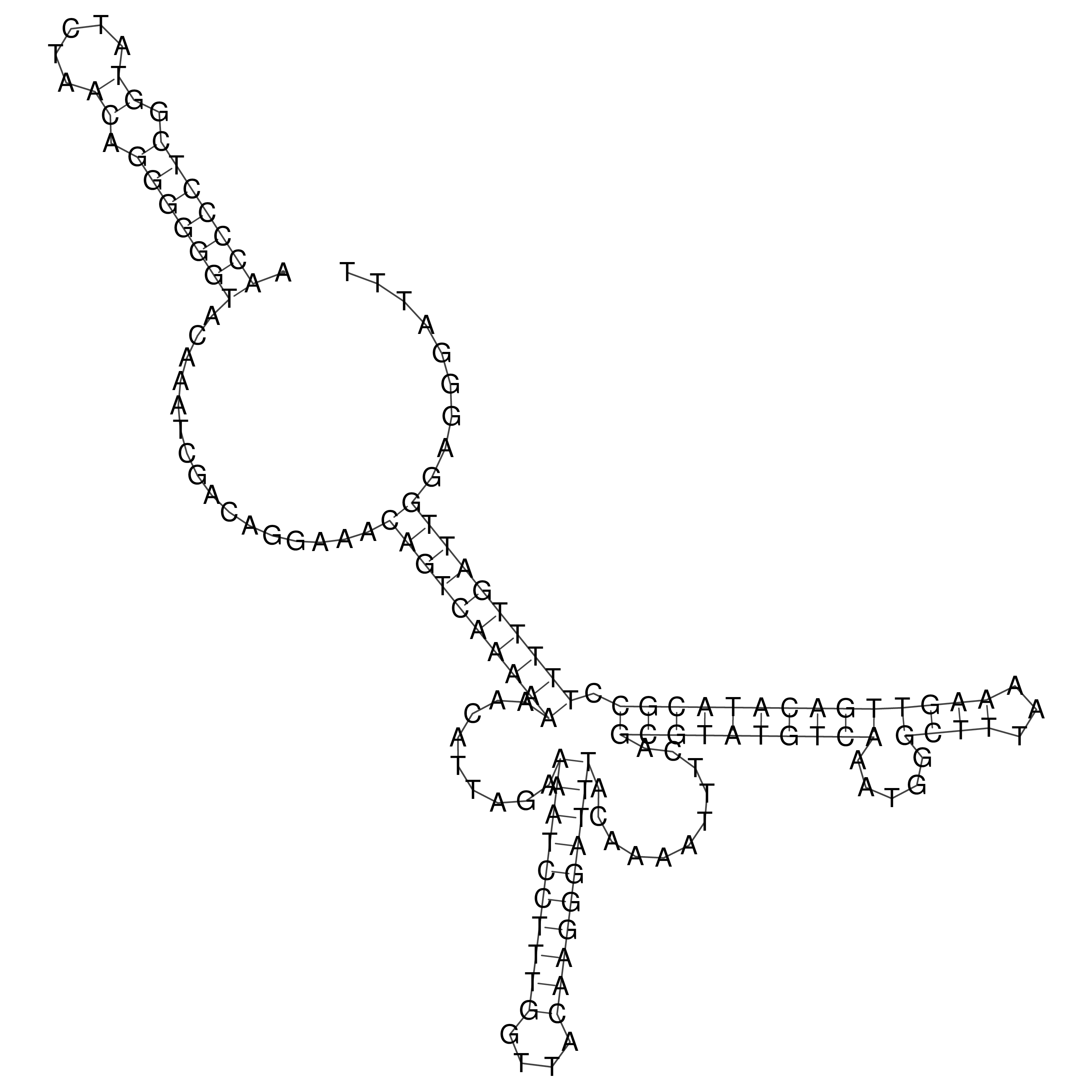
Relaxase
| ID | 266 | GenBank | ADH43079 |
| Name | Relaxase_Tn6079 |
UniProt ID | D7PTR9 |
| Length | 329 a.a. | PDB ID | |
| Note | Tn916 orf20 protein | ||
Relaxase protein sequence
Download Length: 329 a.a. Molecular weight: 39099.72 Da Isoelectric Point: 7.0754
>ADH43079.1 Tn916 orf20 protein [uncultured bacterium MID12]
MLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGRGC
RQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRSGE
LVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLLVY
DNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAP
TLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGRGC
RQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRSGE
LVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLLVY
DNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAP
TLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | D7PTR9 |
T4CP
| ID | 266 | GenBank | ADH43077 |
| Name | T4CP_Tn6079 |
UniProt ID | D7PTR7 |
| Length | 463 a.a. | PDB ID | _ |
| Note | Tn916-like orf21 protein | ||
T4CP protein sequence
Download Length: 463 a.a. Molecular weight: 53479.10 Da Isoelectric Point: 7.0591
>ADH43077.1 Tn916-like orf21 protein [uncultured bacterium MID12]
MRMIWNKGHRIRASDKHLVYHFSIGMLLFIFVTVLLLLNMKQLMCTDWEHFSLLENGLTLSPYNFITIGI
ATGVCALVAFGYYRFCYDGFKKLLHRQKLARMILENKWYEADTVQDSGFFTDLQSRSREKIVWFPKIYYQ
MEKGLLHIRCEITLGKYQDQLLRLEDKLESGLYCELTDKTLHDGYIEYTLLYDMIANRITIDEVRAENGC
LKLMKNLVWEYDALPHALIAGGTGGGKTYFLLTLIEALLHTNAVLYILDPKNSDLADLGTVMPNVYHTKE
EMIDCVNAFYEGMVQRSEEMKRHPNYKTGENYAYLGLPPCFLIFDEYVAFFEMLGTKESVSLLSQLKKIV
MLGRQAGYFLIVACQRPDAKYFSDGIRDNFNFRVGLGRISELGYGMLFGSDVKKQFFQKRIKGRGYCDVG
TSVISEFYTPLVPKGHDFLQTIGRLAQERQAMPEQQGKEDVIE
MRMIWNKGHRIRASDKHLVYHFSIGMLLFIFVTVLLLLNMKQLMCTDWEHFSLLENGLTLSPYNFITIGI
ATGVCALVAFGYYRFCYDGFKKLLHRQKLARMILENKWYEADTVQDSGFFTDLQSRSREKIVWFPKIYYQ
MEKGLLHIRCEITLGKYQDQLLRLEDKLESGLYCELTDKTLHDGYIEYTLLYDMIANRITIDEVRAENGC
LKLMKNLVWEYDALPHALIAGGTGGGKTYFLLTLIEALLHTNAVLYILDPKNSDLADLGTVMPNVYHTKE
EMIDCVNAFYEGMVQRSEEMKRHPNYKTGENYAYLGLPPCFLIFDEYVAFFEMLGTKESVSLLSQLKKIV
MLGRQAGYFLIVACQRPDAKYFSDGIRDNFNFRVGLGRISELGYGMLFGSDVKKQFFQKRIKGRGYCDVG
TSVISEFYTPLVPKGHDFLQTIGRLAQERQAMPEQQGKEDVIE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | D7PTR7 |
Host bacterium
| ID | 57 | Element type | Transposon |
| Element name | Tn6079 | GenBank | GU951538 |
| Element size | 45360 bp | Coordinate of oriT [Strand] | 7612..7763 [+] |
| Host bacterium | Uncultured bacterium MID12 | Coordinate of element | 462..28872 |
Cargo genes
| Drug resistance gene | tetracycline and erythromycin resistanc |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |