Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200050 |
| Name | oriT_ICEFal35896-1 |
| Organism | Filifactor alocis ATCC 35896 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | CP002390 (1262384..1262597 [-], 214 nt) |
| oriT length | 214 nt |
| IRs (inverted repeats) | 66..76, 80..89 (ACCCCCCGTAT..ACAGGGGGGT) 125..134, 139..148 ( AAATTCTTTG..CAAGGAGTTT) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TGG|T |
| Note | blastn alignment with the oriT_Tn916 |
oriT sequence
Download Length: 214 nt
>oriT_ICEFal35896-1
AAGCGAAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCGATAGCCACGCCAGCACCTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAATAGCAGAAAATTCTTTGGTTACAAGGAGTTTAGAAAATTTCGTGGTATGTCAAATGAGCTTCAATAGTTGACATACCTTTTTTAATTGGAGGGATTT
AAGCGAAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCGATAGCCACGCCAGCACCTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAATAGCAGAAAATTCTTTGGTTACAAGGAGTTTAGAAAATTTCGTGGTATGTCAAATGAGCTTCAATAGTTGACATACCTTTTTTAATTGGAGGGATTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file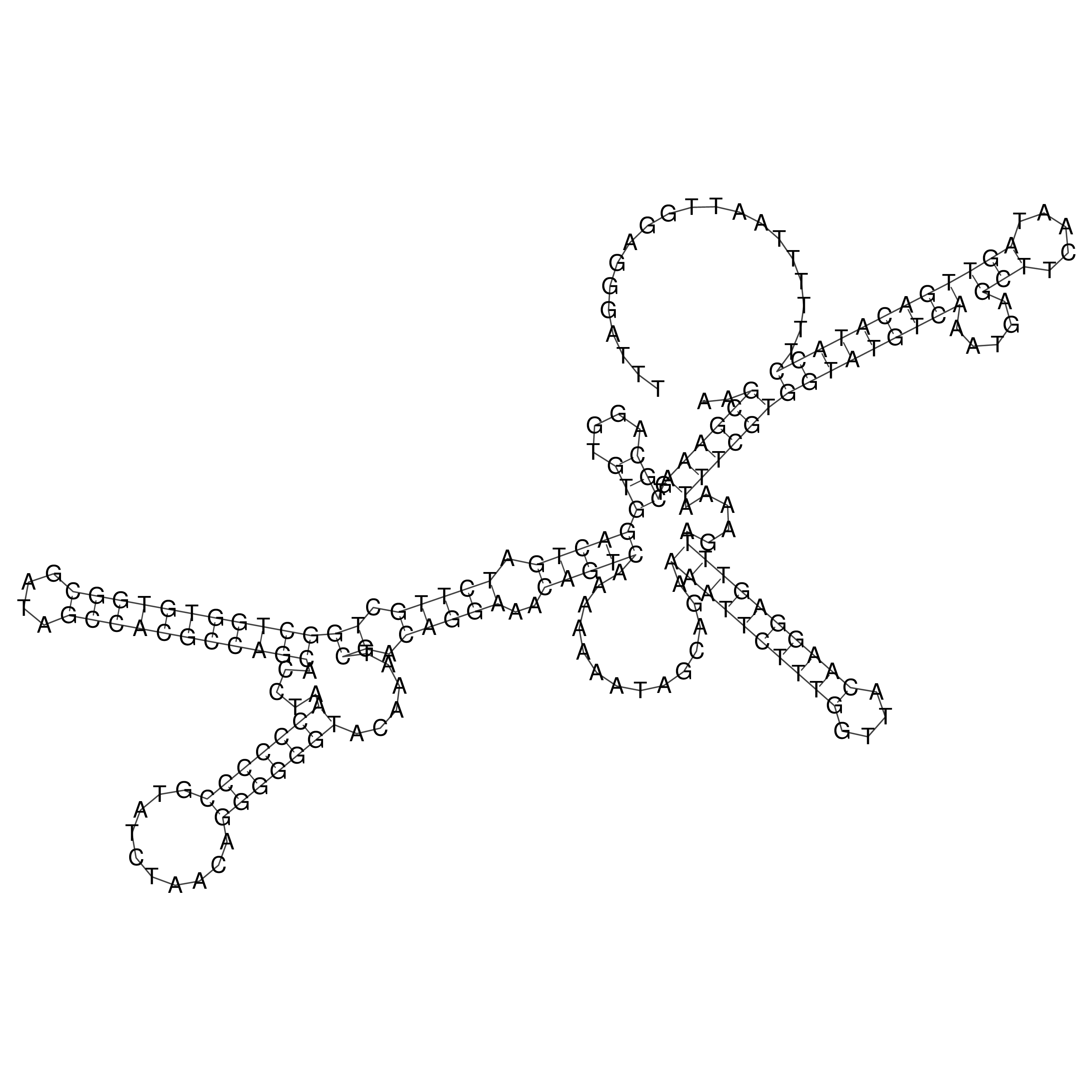
Relaxase
| ID | 264 | GenBank | EFE28724 |
| Name | Relaxase_ICEFal35896-1 |
UniProt ID | _ |
| Length | 391 a.a. | PDB ID | |
| Note | DNA-binding helix-turn-helix protein | ||
Relaxase protein sequence
Download Length: 391 a.a. Molecular weight: 46169.75 Da Isoelectric Point: 6.5913
>EFE28724.1 DNA-binding helix-turn-helix protein [Filifactor alocis ATCC 35896]
MIQHLKEKRLAYGLSQNRLAIATGITRQYLSDIETGKVKPSDELQQSLWETLERFNPDAPLEMLFDYVRI
RFPTMDVQHVVEEILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHEIDKGVLVELKGRGCRQFESYLL
AQQRSWYEFFMDALVAGGVMKRLDLAINDKTGILNIPTLTEKCLQEECISVFRSFKSYRSGELVRKDEKE
CMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLLVYDNPEHTAF
KIINRYIRFVDKDDSKARSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAPTLKVAITL
DEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MIQHLKEKRLAYGLSQNRLAIATGITRQYLSDIETGKVKPSDELQQSLWETLERFNPDAPLEMLFDYVRI
RFPTMDVQHVVEEILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHEIDKGVLVELKGRGCRQFESYLL
AQQRSWYEFFMDALVAGGVMKRLDLAINDKTGILNIPTLTEKCLQEECISVFRSFKSYRSGELVRKDEKE
CMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLLVYDNPEHTAF
KIINRYIRFVDKDDSKARSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAPTLKVAITL
DEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 264 | GenBank | EFE28725 |
| Name | T4CP_ICEFal35896-1 |
UniProt ID | _ |
| Length | 464 a.a. | PDB ID | _ |
| Note | FtsK/SpoIIIE family protein | ||
T4CP protein sequence
Download Length: 464 a.a. Molecular weight: 53690.45 Da Isoelectric Point: 8.2463
>EFE28725.1 FtsK/SpoIIIE family protein [Filifactor alocis ATCC 35896]
MKQFGIRGKRIRPSDKDLVFHFTVASLLPIFLLVIGLFHVKTIQQINWQDFNLSQVDKIHIPYLVISVSV
AILVCLLVAFLFKRYRYDTVKQLYHRQKLAKMVLENKWYESEQVKTDGFFKDSSSRTKEKITYFPKIFYR
LKDGLIQMRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVQAKDGK
LRLMENVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLYTDSKLYILDPKNADLADLGSVMGNVYYRKE
DMLSCIDRFYDEMMARSEAMKEMENYKTGENYAYLGLPANFLIFDEYVAFMEMLGNKENTAVLNKLKQIV
MLGRQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKQIKGRGYVDVG
TSVISEFYTPLVPKGHDFLEEIKKLTNSRQSTQATCEAKVAGVD
MKQFGIRGKRIRPSDKDLVFHFTVASLLPIFLLVIGLFHVKTIQQINWQDFNLSQVDKIHIPYLVISVSV
AILVCLLVAFLFKRYRYDTVKQLYHRQKLAKMVLENKWYESEQVKTDGFFKDSSSRTKEKITYFPKIFYR
LKDGLIQMRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVQAKDGK
LRLMENVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLYTDSKLYILDPKNADLADLGSVMGNVYYRKE
DMLSCIDRFYDEMMARSEAMKEMENYKTGENYAYLGLPANFLIFDEYVAFMEMLGNKENTAVLNKLKQIV
MLGRQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKQIKGRGYVDVG
TSVISEFYTPLVPKGHDFLEEIKKLTNSRQSTQATCEAKVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1253369..1264722
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HMPREF0389_00631 | 1248550..1248753 | - | 204 | EFE28712 | DNA binding domain, excisionase family | - |
| HMPREF0389_00632 | 1249215..1249445 | - | 231 | EFE28713 | hypothetical protein | - |
| HMPREF0389_00633 | 1249442..1249867 | - | 426 | EFE28714 | RNA polymerase sigma factor, sigma-70 family | - |
| HMPREF0389_00634 | 1250378..1250731 | + | 354 | EFE28715 | DNA-binding helix-turn-helix protein | - |
| HMPREF0389_00635 | 1251073..1252992 | - | 1920 | EFE28716 | putative translation elongation factor G | - |
| HMPREF0389_00636 | 1253369..1254283 | - | 915 | EFE28717 | hypothetical protein | orf13 |
| HMPREF0389_00637 | 1254298..1255299 | - | 1002 | EFE28718 | NlpC/P60 family protein | orf14 |
| HMPREF0389_00638 | 1255296..1257473 | - | 2178 | EFE28719 | hypothetical protein | orf15 |
| HMPREF0389_00639 | 1257476..1259923 | - | 2448 | EFE28720 | hypothetical protein | virb4 |
| HMPREF0389_00640 | 1259907..1260338 | - | 432 | EFE28721 | hypothetical protein | orf17a |
| HMPREF0389_00641 | 1260313..1260810 | - | 498 | EFE28722 | antirestriction protein ArdA | - |
| HMPREF0389_00642 | 1260927..1261148 | - | 222 | EFE28723 | hypothetical protein | orf19 |
| HMPREF0389_00643 | 1261191..1262366 | - | 1176 | EFE28724 | DNA-binding helix-turn-helix protein | - |
| HMPREF0389_00644 | 1262572..1263966 | - | 1395 | EFE28725 | FtsK/SpoIIIE family protein | virb4 |
| HMPREF0389_00645 | 1264006..1264389 | - | 384 | EFE28726 | hypothetical protein | orf23 |
| HMPREF0389_00646 | 1264408..1264722 | - | 315 | EFE28727 | hypothetical protein | orf23 |
| HMPREF0389_00647 | 1265051..1266049 | - | 999 | EFE28728 | ABC transporter, ATP-binding protein | - |
| HMPREF0389_00648 | 1266046..1266774 | - | 729 | EFE28729 | cobalt transport protein | - |
| HMPREF0389_00649 | 1266774..1267364 | - | 591 | EFE28730 | conserved hypothetical protein TIGR02185 | - |
| HMPREF0389_00650 | 1267390..1269114 | - | 1725 | EFE28731 | ABC transporter, ATP-binding protein | - |
Host bacterium
| ID | 55 | Element type | |
| Element name | ICEFal35896-1 | GenBank | CP002390 |
| Element size | 1931012 bp | Coordinate of oriT [Strand] | 1262384..1262597 [-] |
| Host bacterium | Filifactor alocis ATCC 35896 | Coordinate of element | 1247077..1265062 |
Cargo genes
| Drug resistance gene | tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |