Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200040 |
| Name | oriT_ICECdiM68-1 |
| Organism | Clostridioides difficile M68 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | NC_017175 (337912..338095 [+], 184 nt) |
| oriT length | 184 nt |
| IRs (inverted repeats) | 1..11, 16..26 (GCTGGTGTGGC..GCCACGCCAGC) 92..103, 108..119 ( GAAAATCCTTTG..CAAGGGATTTAC) |
| Location of nic site | 104..105 |
| Conserved sequence flanking the nic site |
TGG|T |
| Note | blastn alignment with the oriT_Tn916 |
oriT sequence
Download Length: 184 nt
>oriT_ICECdiM68-1
GCTGGTGTGGCGTAAGCCACGCCAGCAGGTAAAACCCCTCGGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGATTGGAGGGATTT
GCTGGTGTGGCGTAAGCCACGCCAGCAGGTAAAACCCCTCGGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGATTGGAGGGATTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file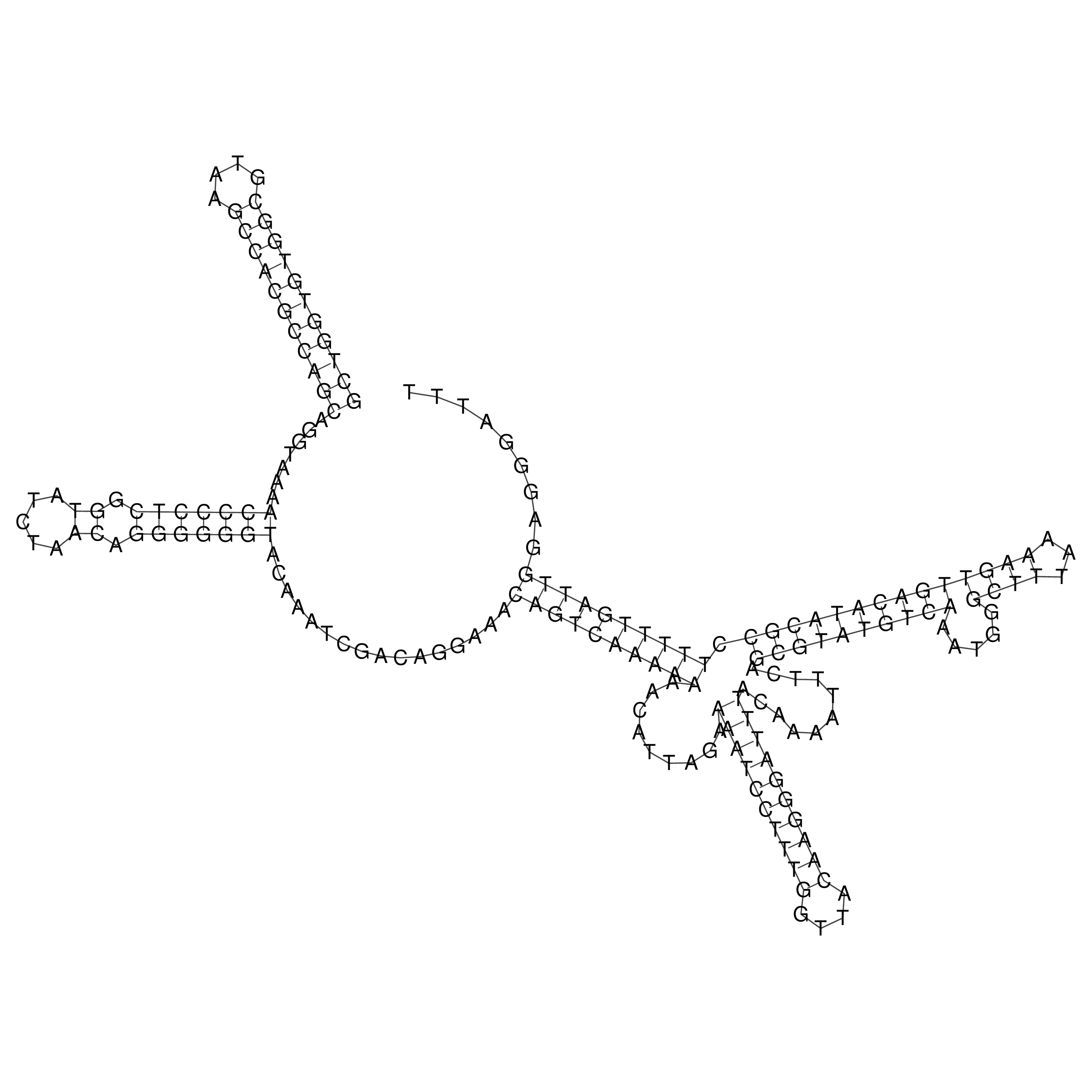
Relaxase
| ID | 254 | GenBank | WP_000398284 |
| Name | Relaxase_ICECdiM68-1 |
UniProt ID | _ |
| Length | 401 a.a. | PDB ID | |
| Note | _ | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
>WP_000398284.1 MULTISPECIES: MobT family relaxase [Bacteria]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 254 | GenBank | WP_044593066 |
| Name | T4CP_ICECdiM68-1 |
UniProt ID | _ |
| Length | 464 a.a. | PDB ID | _ |
| Note | cell division protein FtsK | ||
T4CP protein sequence
Download Length: 464 a.a. Molecular weight: 53309.79 Da Isoelectric Point: 6.8996
>WP_044593066.1 MULTISPECIES: FtsK/SpoIIIE domain-containing protein [Clostridia]
MRMIWNKGHRIRASDKHLVYHFSIGTLLFVFVAVLLLLNSKQLMCTDWEHFSLLENGLTLSPYNFITILI
ATGVCALVAFLYYRFCYDSFKKLLHRQKLARMVLENKWYEADTVQDSIFFTDLQSRSREKIVWFPKIYYQ
MEKGLLHIRCEITLGKYQDQLLRLEDKLESGLYCELTDKTLHDGYIEYTLLYDMIANRITIDEVRAENGC
LRLMKNLVWEYDALPHALIAGGTGGGKTYFLLTLIEALLHTNAVLYILDPKNADLADLGTVMGNVYHTKE
EMIDCVNAFYEGMVQRSEEMKRHPDYKTGENYAYLGLPPCFLIFDEYVAFFEMLGTKESVSLLSQLKKIV
MLGRQAGYFLIVACQRPDAKYFSDGIRDNFNFRVGLGRISELGYGMLFGSDVKKQFFQKRIKGRGYCDVG
TSVISEFYTPLVPKGHDFLQTIGPLAQARQDVTATCEAKGDGTD
MRMIWNKGHRIRASDKHLVYHFSIGTLLFVFVAVLLLLNSKQLMCTDWEHFSLLENGLTLSPYNFITILI
ATGVCALVAFLYYRFCYDSFKKLLHRQKLARMVLENKWYEADTVQDSIFFTDLQSRSREKIVWFPKIYYQ
MEKGLLHIRCEITLGKYQDQLLRLEDKLESGLYCELTDKTLHDGYIEYTLLYDMIANRITIDEVRAENGC
LRLMKNLVWEYDALPHALIAGGTGGGKTYFLLTLIEALLHTNAVLYILDPKNADLADLGTVMGNVYHTKE
EMIDCVNAFYEGMVQRSEEMKRHPDYKTGENYAYLGLPPCFLIFDEYVAFFEMLGTKESVSLLSQLKKIV
MLGRQAGYFLIVACQRPDAKYFSDGIRDNFNFRVGLGRISELGYGMLFGSDVKKQFFQKRIKGRGYCDVG
TSVISEFYTPLVPKGHDFLQTIGPLAQARQDVTATCEAKGDGTD
Protein domains
No domain identified.
Protein structure
No available structure.
Host bacterium
| ID | 45 | Element type | Transposon |
| Element name | ICECdiM68-1 | GenBank | NC_017175 |
| Element size | 4308325 bp | Coordinate of oriT [Strand] | 337912..338095 [+] |
| Host bacterium | Clostridioides difficile M68 | Coordinate of element | 334783..353473 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |