Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200033 |
| Name | oriT_ICESauST398-1 |
| Organism | Staphylococcus aureus subsp. aureus ST398 |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | AM990992 (1013148..1013362 [-], 215 nt) |
| oriT length | 215 nt |
| IRs (inverted repeats) | 61..76, 80..95 (ACTTAACCCCCCGTAT..ACAGGGGGGTACAAAT) 123..134, 139..150 (GAAAATCCTTTG..CAAGGGATTTAC) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TGG|T |
| Note | blastn alignment with the oriT_Tn916 |
oriT sequence
Download Length: 215 nt
>oriT_ICESauST398-1
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGATTGGAGGGATTT
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGATTGGAGGGATTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file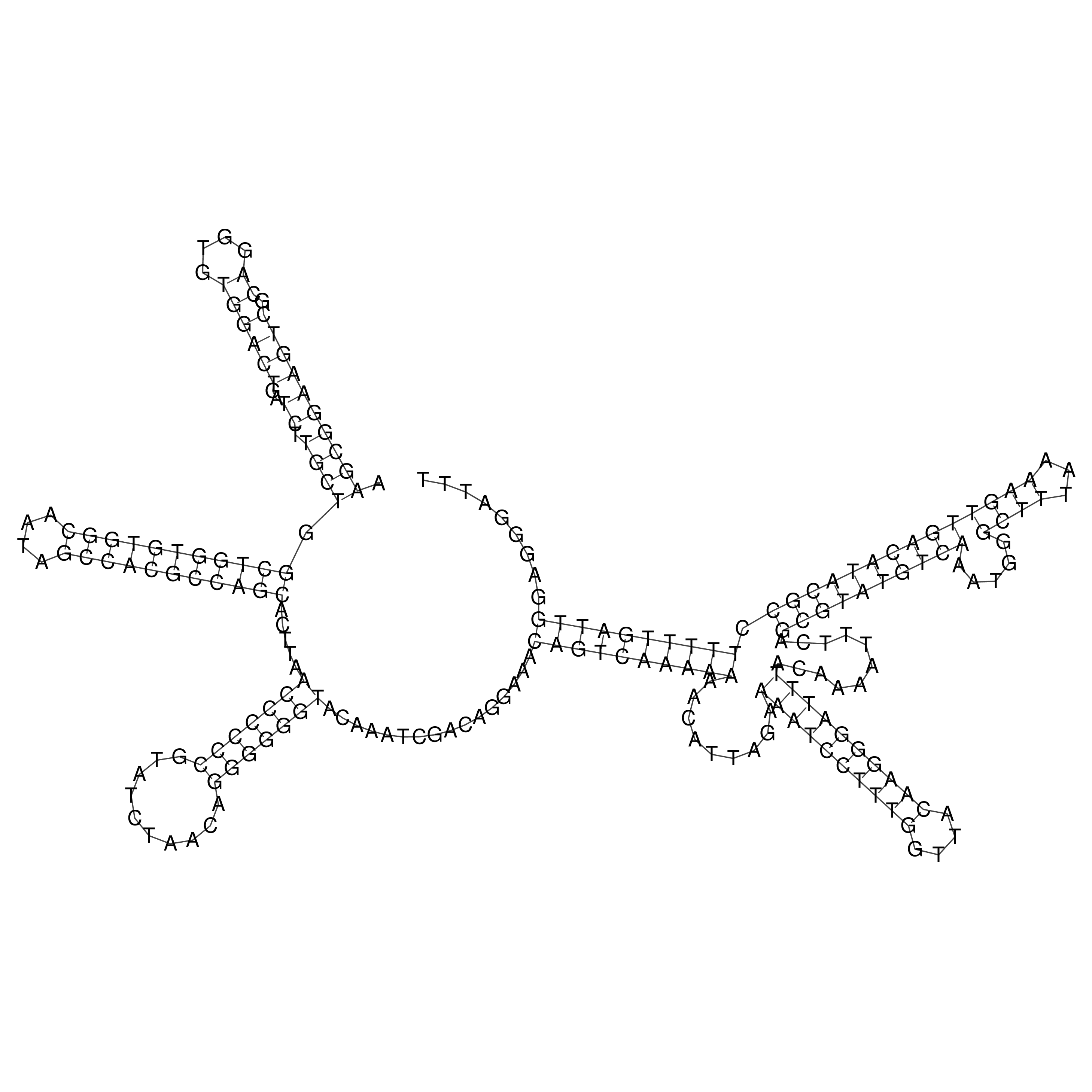
Relaxase
| ID | 247 | GenBank | CAQ49392 |
| Name | Relaxase_ICESauST398-1 |
UniProt ID | D2N5U4 |
| Length | 401 a.a. | PDB ID | |
| Note | transcriptional regulator, Cro/CI family | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.06 Da Isoelectric Point: 6.3243
>CAQ49392.1 transcriptional regulator, Cro/CI family [Staphylococcus aureus subsp. aureus ST398]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLQLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLQLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 247 | GenBank | CAQ49393 |
| Name | T4CP_ICESauST398-1 |
UniProt ID | D2N5U5 |
| Length | 461 a.a. | PDB ID | _ |
| Note | ftsk/spoiiie family protein | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>CAQ49393.1 ftsk/spoiiie family protein [Staphylococcus aureus subsp. aureus ST398]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1004057..1015305
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SAPIG0954 | 999244..999447 | - | 204 | CAQ49381 | conserved domain protein | - |
| SAPIG0955 | 1000135..1000557 | - | 423 | CAQ49382 | sigma-70, region 4 family | - |
| SAPIG0956 | 1001062..1001415 | + | 354 | CAQ49383 | transcriptional regulator, putative | - |
| SAPIG0957 | 1001761..1003680 | - | 1920 | CAQ49384 | TetM protein | - |
| SAPIG0958 | 1004057..1004989 | - | 933 | CAQ49385 | conjugative transposon protein | orf13 |
| SAPIG0959 | 1004986..1005987 | - | 1002 | CAQ49386 | NLP/P60 family protein | orf14 |
| SAPIG0960 | 1005984..1008161 | - | 2178 | CAQ49387 | conjugative transposon membrane protein | orf15 |
| SAPIG0961 | 1008164..1010611 | - | 2448 | CAQ49388 | conjugative transposon protein | virb4 |
| SAPIG0962 | 1010595..1010987 | - | 393 | CAQ49389 | conjugative transposon membrane protein | orf17a |
| SAPIG0963 | 1011076..1011573 | - | 498 | CAQ49390 | conjugative transposon protein | - |
| SAPIG0964 | 1011690..1011911 | - | 222 | CAQ49391 | conserved domain protein | orf19 |
| SAPIG0965 | 1011954..1013159 | - | 1206 | CAQ49392 | transcriptional regulator, Cro/CI family | - |
| SAPIG0966 | 1013337..1014722 | - | 1386 | CAQ49393 | ftsk/spoiiie family protein | virb4 |
| SAPIG0967 | 1014751..1015137 | - | 387 | CAQ49394 | conjugative transposon protein | orf23 |
| SAPIG0968 | 1015153..1015305 | - | 153 | CAQ49395 | conjugative transposon protein | orf23 |
| SAPIG0969 | 1015836..1016147 | - | 312 | CAQ49396 | transposase | - |
| SAPIG0970 | 1017038..1018354 | - | 1317 | CAQ49397 | CoA-disulfide reductase | - |
| SAPIG0971 | 1018406..1019230 | - | 825 | CAQ49398 | hydrolase | - |
| SAPIG0972 | 1019344..1019652 | + | 309 | CAQ49399 | N-6 Adenine-specific DNA methylase YitW | - |
Host bacterium
| ID | 38 | Element type | Transposon |
| Element name | ICESauST398-1 | GenBank | AM990992 |
| Element size | 2872582 bp | Coordinate of oriT [Strand] | 1013148..1013362 [-] |
| Host bacterium | Staphylococcus aureus subsp. aureus ST398 | Coordinate of element | 997771..1015802 |
Cargo genes
| Drug resistance gene | tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |