Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200023 |
| Name | oriT_CTn6002 |
| Organism | Streptococcus cristatus |
| Sequence Completeness | intact |
| NCBI accession of oriT (coordinates [strand]) | AY898750 (2441..2655 [+], 215 nt) |
| oriT length | 215 nt |
| IRs (inverted repeats) | 61..76, 80..95 (ACTTAACCCCCCGTAT..ACAGGGGGGTACAAAT) 123..134, 139..150 (GAAAATCCTTTG..CAAGGGATTTAC) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TGG|T |
| Note | blastn alignment with the oriT_Tn916 |
oriT sequence
Download Length: 215 nt
>oriT_CTn6002
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGATTGGAGGGATTT
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGATTGGAGGGATTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file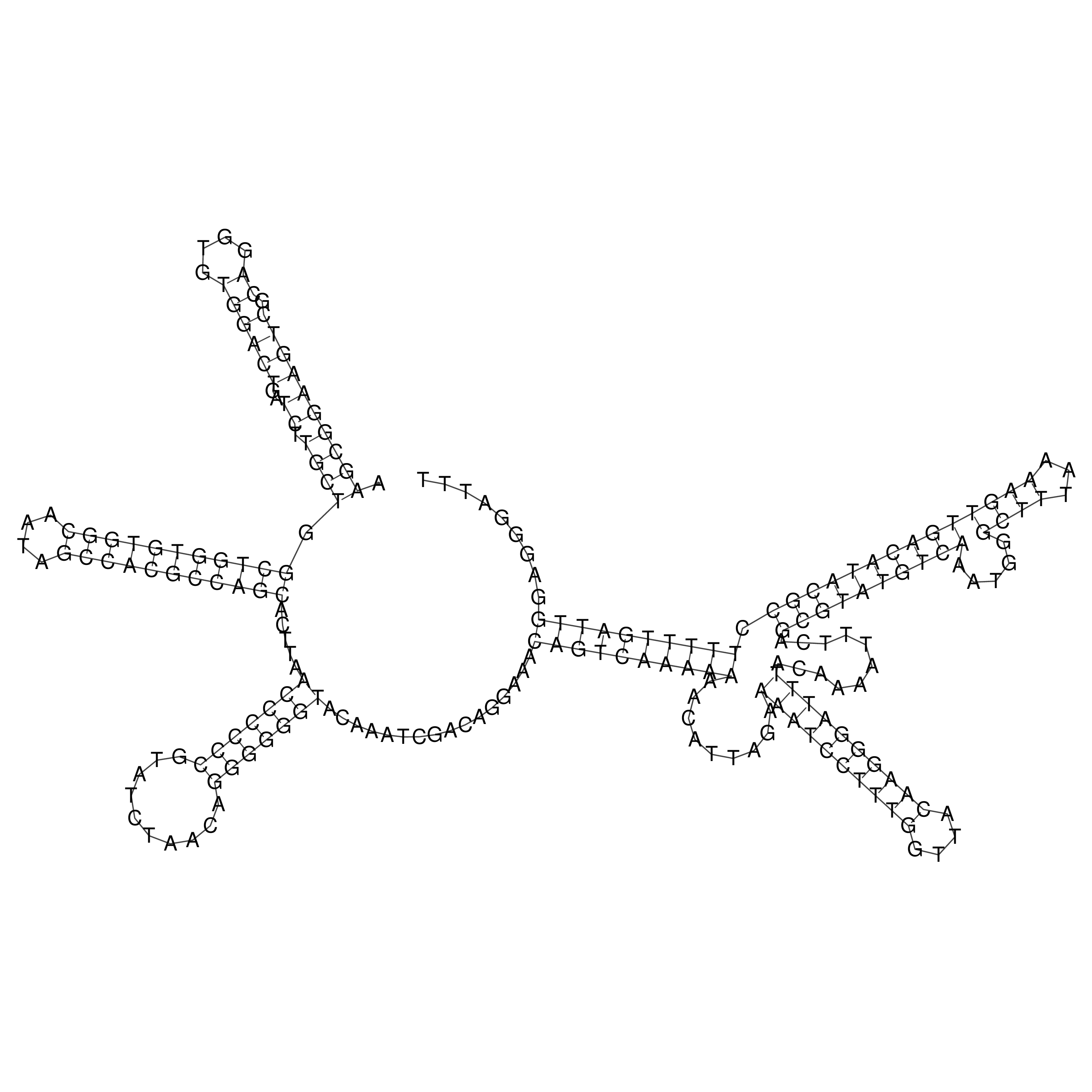
Relaxase
| ID | 237 | GenBank | AAY63930 |
| Name | ORF20_CTn6002 |
UniProt ID | Q3KU49 |
| Length | 400 a.a. | PDB ID | |
| Note | ORF20F; similar to ORF20 in Tn916, YP_133675, but extended by 72 amino acids | ||
Relaxase protein sequence
Download Length: 400 a.a. Molecular weight: 47341.51 Da Isoelectric Point: 8.8918
>AAY63930.1 hypothetical protein [Streptococcus cristatus]
MLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGRGC
RQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRSGE
LVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLLVY
DNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAP
TLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKKGYLSTIPVDRYPKKDIMGDKT
VRVRADLHHIIKIETAKNGGNVKEVMEIRLRSKLKSVLIVHYLKILYNRN
MLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGRGC
RQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRSGE
LVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLLVY
DNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQVAP
TLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKKGYLSTIPVDRYPKKDIMGDKT
VRVRADLHHIIKIETAKNGGNVKEVMEIRLRSKLKSVLIVHYLKILYNRN
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q3KU49 |
T4CP
| ID | 237 | GenBank | AAY63929 |
| Name | ORF21_CTn6002 |
UniProt ID | Q3KU50 |
| Length | 461 a.a. | PDB ID | _ |
| Note | ORF21; similar to ORF21 in Tn916, YP_133674, and DNA segregation ATPase, FtsK/SpoIIIE family protein, ZP_00285890 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>AAY63929.1 hypothetical protein [Streptococcus cristatus]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q3KU50 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 336..14594
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| Locus_0 | 194..313 | + | 120 | AAY63926 | hypothetical protein | - |
| Locus_1 | 336..650 | + | 315 | AAY63927 | hypothetical protein | orf23 |
| Locus_2 | 666..1052 | + | 387 | AAY63928 | hypothetical protein | orf23 |
| Locus_3 | 1081..2466 | + | 1386 | AAY63929 | hypothetical protein | virb4 |
| Locus_4 | 2860..4062 | + | 1203 | AAY63930 | hypothetical protein | - |
| Locus_5 | 3895..4062 | + | 168 | ABL76174 | MLS leader peptide | - |
| Locus_6 | 4111..4194 | + | 84 | AAY63931 | MLS leader peptide | - |
| Locus_7 | 4319..5056 | + | 738 | AAY63932 | MLS methylase | - |
| Locus_8 | 5001..5192 | + | 192 | AAY63933 | hypothetical protein | - |
| Locus_9 | 5313..6560 | - | 1248 | AAY63934 | hypothetical protein | - |
| Locus_10 | 6740..6961 | + | 222 | AAY63935 | hypothetical protein | orf19 |
| Locus_11 | 7078..7575 | + | 498 | AAY63936 | hypothetical protein | - |
| Locus_12 | 7550..8056 | + | 507 | AAY63937 | hypothetical protein | orf17a |
| Locus_13 | 8040..10487 | + | 2448 | AAY63938 | hypothetical protein | virb4 |
| Locus_14 | 10490..12667 | + | 2178 | AAY63939 | hypothetical protein | orf15 |
| Locus_15 | 12664..13449 | + | 786 | AAY63940 | hypothetical protein | orf14 |
| Locus_16 | 13662..14594 | + | 933 | AAY63941 | hypothetical protein | orf13 |
| Locus_17 | 14869..14955 | + | 87 | AAY63942 | hypothetical protein | - |
| Locus_18 | 14971..16890 | + | 1920 | AAY63943 | tetracycline resistance protein TetM | - |
| Locus_19 | 16988..17176 | + | 189 | AAY63951 | hypothetical protein | - |
| Locus_20 | 17236..17589 | - | 354 | AAY63944 | hypothetical protein | - |
| Locus_21 | 17794..17865 | + | 72 | AAY63945 | hypothetical protein | - |
| Locus_22 | 18043..18516 | + | 474 | AAY63946 | hypothetical protein | - |
| Locus_23 | 18513..18743 | + | 231 | AAY63947 | hypothetical protein | - |
| Locus_24 | 18969..19220 | - | 252 | AAY63948 | hypothetical protein | - |
| Locus_25 | 19204..19407 | + | 204 | AAY63949 | Xis-Tn | - |
Host bacterium
| ID | 28 | Element type | Transposon |
| Element name | CTn6002 | GenBank | AY898750 |
| Element size | 20880 bp | Coordinate of oriT [Strand] | 2441..2655 [+] |
| Host bacterium | Streptococcus cristatus |
Cargo genes
| Drug resistance gene | erm(B), tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |