Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 200006 |
| Name | oriT_ICEBs1 |
| Organism | Bacillus subtilis subsp. subtilis str. 168 |
| Sequence Completeness | core |
| NCBI accession of oriT (coordinates [strand]) | AL009126 (534790..534813 [+], 24 nt) |
| oriT length | 24 nt |
| IRs (inverted repeats) | 1..7, 18..24 (ACCCCCC..GGGGGGT) |
| Location of nic site | 13..14 |
| Conserved sequence flanking the nic site |
_ |
| Note | With the help of relaxase YdcR(NicK)_ICEBs1, a helicase processivity factor (HelP, formerly YdcP) can associate with the oriT_ICEBs1 in vivo [PMID:23326247]. |
oriT sequence
Download Length: 24 nt
ACCCCCCCACGCTAACAGGGGGGT
Visualization of oriT structure (The oriT was characterized experimentally)
oriT secondary structure
Predicted by RNAfold.
Download structure file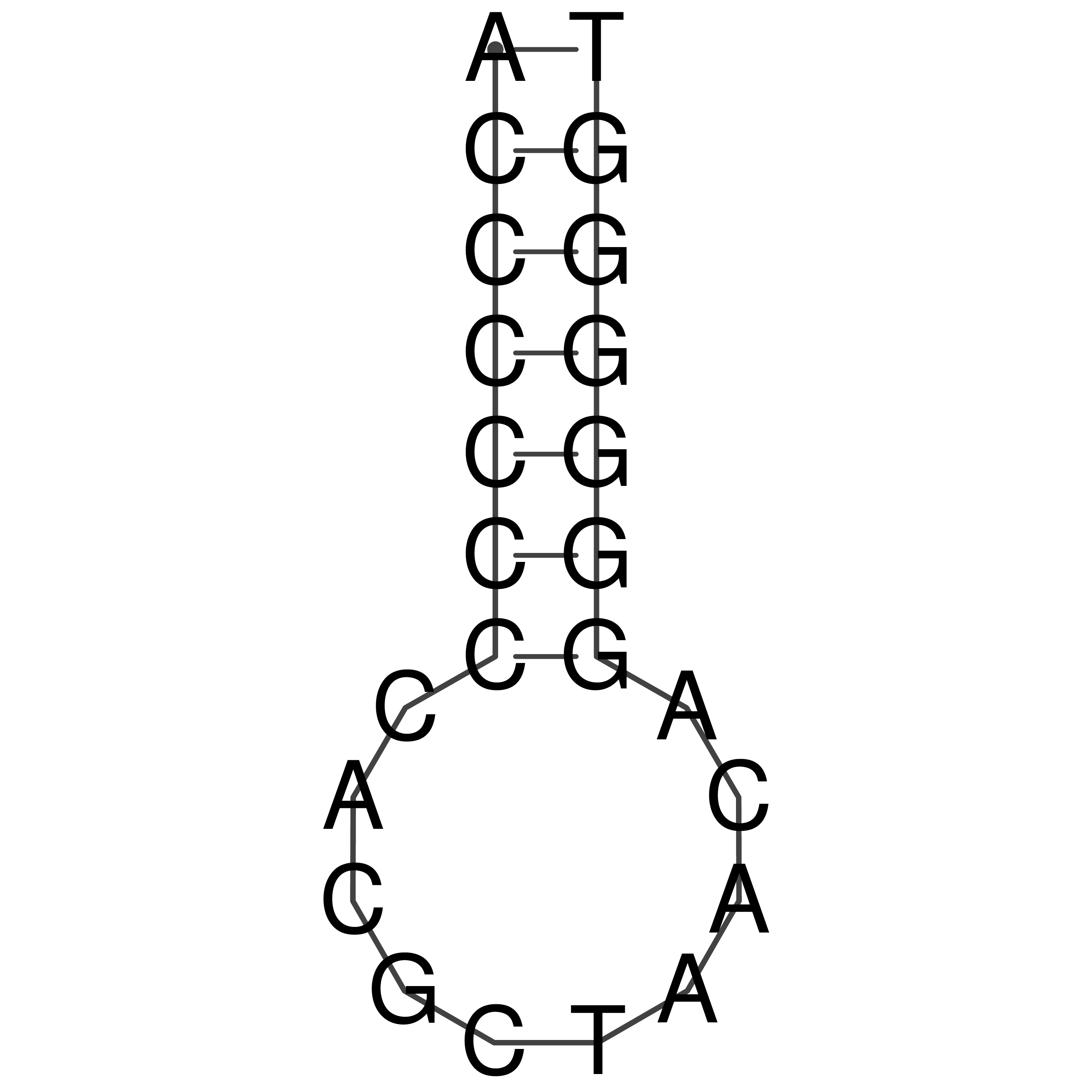
Reference
[1] Lee CA et al. (2012) The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J Bacteriol. 194(12):3165-72. [PMID:22505685]
[2] Lee CA et al. (2010) Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol. 75(2):268-79. [PMID:19943900]
[3] Lee CA et al. (2007) Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol. 189(20):7254-61. [PMID:17693500]
Relaxase
| ID | 220 | GenBank | CAB12294 |
| Name | YdcR(NicK)_ICEBs1 |
UniProt ID | P96635 |
| Length | 352 a.a. | PDB ID | |
| Note | With the help of relaxase YdcR(NicK)_ICEBs1, a helicase processivity factor (HelP, formerly YdcP) can associate with the oriT_ICEBs1 in vivo [PMID:23326247]. | ||
Relaxase protein sequence
Download Length: 352 a.a. Molecular weight: 40943.59 Da Isoelectric Point: 5.1091
MDELKQPPHANRGVVIVKEKNEAVESPLVSMVDYIRVSFKTHDVDRIIEEVLHLSKDFMTEKQSGFYGYV
GTYELDYIKVFYSAPDDNRGVLIEMSGQGCRQFESFLECRKKTWYDFFQDCMQQGGSFTRFDLAIDDKKT
YFSIPELLKKAQKGECISRFRKSDFNGSFDLSDGITGGTTIYFGSKKSEAYLCFYEKNYEQAEKYNIPLE
ELGDWNRYELRLKNERAQVAIDALLKTKDLTLIAMQIINNYVRFVDADENITREHWKTSLFWSDFIGDVG
RLPLYVKPQKDFYQKSRNWLRNSCAPTMKMVLEADEHLGKTDLSDMIAEAELADKHKKMLDVYMADVADM
VV
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | P96635 |
Reference
[1] Lee CA et al. (2012) The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J Bacteriol. 194(12):3165-72. [PMID:22505685]
[2] Lee CA et al. (2010) Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol. 75(2):268-79. [PMID:19943900]
[3] Lee CA et al. (2007) Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol. 189(20):7254-61. [PMID:17693500]
T4CP
| ID | 220 | GenBank | CAB12293 |
| Name | YdcQ_ICEBs1 |
UniProt ID | P96634 |
| Length | 480 a.a. | PDB ID | _ |
| Note | putative coupling protein | ||
T4CP protein sequence
Download Length: 480 a.a. Molecular weight: 54741.88 Da Isoelectric Point: 9.3968
MSDFLNKRFWKYRGKRIRPYMRNNVKLAGAIIFVPVFLLSMFLFWREQLIHFDLSQVIKNFEWNVPLIIK
SVLCSVLIAVGSIVASYFLLFDSYKKILHRQKIAKMIFSNKFYEKENVKVRKIFSNETDSKEKITYFPRM
YYQVKNNHIYIRIAMDMSRFQNRFLDLGKDLENGLFCDLVDKQMEEGFVCFKLLYDVKKNRISIDDAVAE
NGVLPLMKHISWQFDKLPHMLIAGGTGGGKTYFMLTIIKACVGLGADVRILDPKNADLADLEEVLPKKVY
SQKNGILMCLRKSVDGMMERMDEMKQMSNYKTGENYAYLGLKPVFIFFDEYVAFMDLLDMKERNEALSYM
KQLVMLGRQAGYFLVLGAQRPDAKYLADGIRDQFSFRVSLGLMSDTGYGMMFGDVEKAYVNKKETGRGYA
NVGTGSVLEFYSPIVPKGYDFMSSIKNALVGVEGAQATAVASGSVSDQTASGEGVSEANG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | P96634 |
Reference
[1] Lee CA et al. (2007) Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J Bacteriol. 189(20):7254-61. [PMID:17693500]
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 532922..545011
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BSU_04790 (BSU04790) | 528129..528581 | + | 453 | CAB12286 | conserved protein of unknown function | - |
| BSU_04800 (BSU04800) | 529505..530611 | - | 1107 | CAB12287 | ICEBs1 mobile element: integrase | - |
| BSU_04810 (BSU04810) | 530624..531133 | - | 510 | CAB12288 | ICEBs1 mobile element: site-specific protease cleaving ImmR | - |
| BSU_04820 (BSU04820) | 531130..531513 | - | 384 | CAB12289 | ICEBs1 mobile element: transcriptional regulator (Xre family) | - |
| BSU_04830 (BSU04830) | 531787..531981 | + | 195 | CAB12290 | ICEBs1 mobile element: excisionase | - |
| BSU_04839 (BSU04839) | 531978..532238 | + | 261 | CAX52552 | ICEBs1 mobile element: conserved protein of unknown function | - |
| BSU_04840 (BSU04840) | 532292..532552 | + | 261 | CAB12291 | ICEBs1 mobile element: inhibitor of biofilm formation and sporulation | - |
| BSU_04849 (BSU04849) | 532758..532886 | + | 129 | SOX90545 | ICEBs1 mobile element: hypothetical protein | - |
| BSU_04850 (BSU04850) | 532922..533302 | + | 381 | CAB12292 | ICEBs1 mobile element: helicase processivity factor | orf23 |
| BSU_04860 (BSU04860) | 533338..534780 | + | 1443 | CAB12293 | ICEBs1 mobile element: coupling type IV secretion system conjugation protein VirD4 | virb4 |
| BSU_04870 (BSU04870) | 534773..535831 | + | 1059 | CAB12294 | ICEBs1 mobile element: DNA relaxase | - |
| BSU_04880 (BSU04880) | 536096..536365 | + | 270 | CAB12295 | ICEBs1 mobile element: conserved protein of unknown function | - |
| BSU_04890 (BSU04890) | 536404..536670 | + | 267 | CAB12296 | ICEBs1 mobile element: conserved protein of unknown function | - |
| BSU_04900 (BSU04900) | 536687..536995 | + | 309 | CAB12297 | ICEBs1 mobile element: conserved protein of unknown function | - |
| BSU_04910 (BSU04910) | 536985..538049 | + | 1065 | CAB12298 | ICEBs1 mobile element: subunit of the conjugation machinery VirB8 for localization of ConE to the membrane | orf13 |
| BSU_04920 (BSU04920) | 538061..538309 | + | 249 | CAB12299 | ICEBs1 mobile element: subunit of the conjugation machinery | - |
| BSU_04930 (BSU04930) | 538322..538846 | + | 525 | CAB12300 | ICEBs1 mobile element: subunit of the conjugation machinery | orf17b |
| BSU_04940 (BSU04940) | 538734..541229 | + | 2496 | CAB12301 | ICEBs1 mobile element: VirB4-like ATPase | virb4 |
| BSU_04950 (BSU04950) | 541248..541574 | + | 327 | CAB12302 | ICEBs1 mobile element: conserved protein of unknown function | - |
| BSU_04960 (BSU04960) | 541578..544025 | + | 2448 | CAB12303 | ICEBs1 mobile element: VirB6 subunit of the conjugation machinery target of ConJ-mediated exclusion | orf15 |
| BSU_04970 (BSU04970) | 544022..545011 | + | 990 | CAB12304 | ICEBs1 mobile element: two-domain autolysin with N-acetylmuramidase and DL-endopeptidase activity VirB1 | orf14 |
| BSU_04980 (BSU04980) | 545026..545532 | + | 507 | CAB12305 | ICEBs1 mobile element: conserved protein of unknown function | - |
| BSU_04990 (BSU04990) | 545595..545975 | + | 381 | CAB12306 | ICEBs1 mobile element: lipoprotein confering ISEBs1 exclusion specificity | - |
| BSU_05000 (BSU05000) | 546166..546966 | - | 801 | CAB12307 | anti-phage protection factor; ICEBs1 mobile element | - |
| BSU_05010 (BSU05010) | 547306..548481 | + | 1176 | CAB12308 | ICEBs1 mobile element: response regulator aspartate phosphatase | - |
| BSU_05020 (BSU05020) | 548438..548557 | + | 120 | CAB12309 | ICEBs1 mobile element: secreted regulator of the activity of phosphatase RapI | - |
| BSU_05030 (BSU05030) | 548710..549651 | + | 942 | CAB12310 | ICEBs1 mobile element: putative helicase | - |
Host bacterium
| ID | 11 | Element type | ICE (Integrative and conjugative element) |
| Element name | ICEBs1 | GenBank | AL009126 |
| Element size | 4215606 bp | Coordinate of oriT [Strand] | 534790..534813 [+] |
| Host bacterium | Bacillus subtilis subsp. subtilis str. 168 | Coordinate of element | 529423..549932 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIII1 |