Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 122568 |
| Name | oriT1_E8377|3 |
| Organism | Enterococcus faecium isolate E8377 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LR135403 (27904..27933 [-], 30 nt) |
| oriT length | 30 nt |
| IRs (inverted repeats) | 1..8, 13..20 (GGGAGTGT..ACACTCCC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 30 nt
>oriT1_E8377|3
GGGAGTGTAGCCACACTCCCTATGCTTGCT
GGGAGTGTAGCCACACTCCCTATGCTTGCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file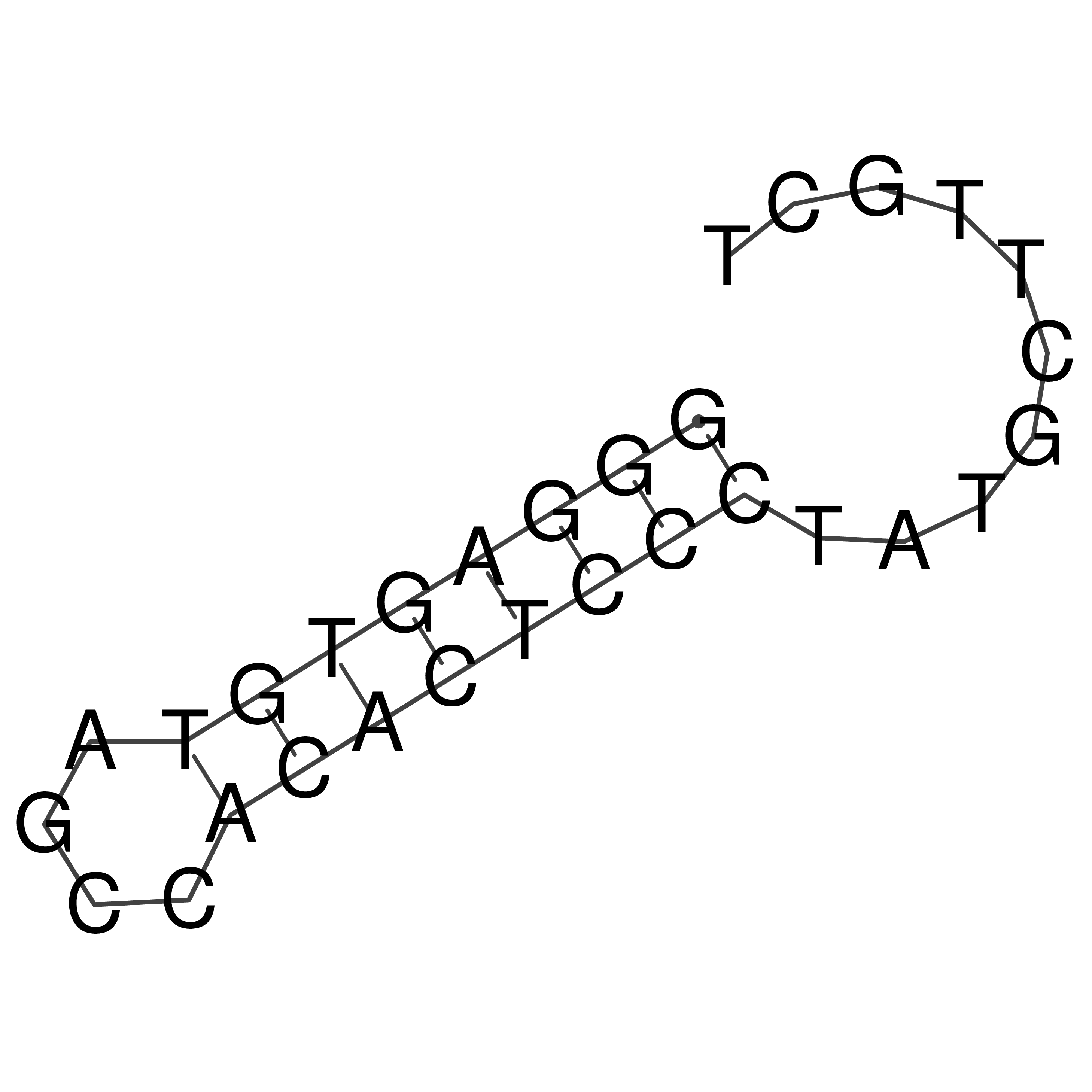
Relaxase
| ID | 14231 | GenBank | WP_002317279 |
| Name | Relaxase_EQB54_RS16630_E8377|3 |
UniProt ID | N2BU79 |
| Length | 442 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 442 a.a. Molecular weight: 50173.57 Da Isoelectric Point: 9.6754
>WP_002317279.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Terrabacteria group]
MAVTKIHPIKSTLKKALDYIENPDKTDGKLLVASFACSYETADIEFEMLLAQAYQKGNNLGHHLIQSFAP
GEATPEQAHEIGKQLAEEVLQGKYPYVLTTHIDKGHVHNHIIFCAVDMVNQRKYISNKQSYAYIRRTSDR
LCKENGLSVVKPGQSKGKSYAEWDAQRKGTSWKAKLKAAIDAAIPQAKDFDDFLRLMQAQGYEIKPGKFI
SFRAPGQERFTRCKTLGEAYTEEAIKERIKGRVITRAPKERKGISLRIDLENNIKAQQSAGYERWAKLHN
LKQAAKTLNFLTEHGIDTYPDLESKVAEITAASDEAAATLKAMEHRLADMAVLIKNISTYKQLWPVAMEY
RNAADKAKFRREHESTLILYEAAAKALKGQGVKKLPDLYALKAEYKRLAEEKERLYEKYGEAKKQMQEYG
IIKQNVDGILRMTPGKEWTQEL
MAVTKIHPIKSTLKKALDYIENPDKTDGKLLVASFACSYETADIEFEMLLAQAYQKGNNLGHHLIQSFAP
GEATPEQAHEIGKQLAEEVLQGKYPYVLTTHIDKGHVHNHIIFCAVDMVNQRKYISNKQSYAYIRRTSDR
LCKENGLSVVKPGQSKGKSYAEWDAQRKGTSWKAKLKAAIDAAIPQAKDFDDFLRLMQAQGYEIKPGKFI
SFRAPGQERFTRCKTLGEAYTEEAIKERIKGRVITRAPKERKGISLRIDLENNIKAQQSAGYERWAKLHN
LKQAAKTLNFLTEHGIDTYPDLESKVAEITAASDEAAATLKAMEHRLADMAVLIKNISTYKQLWPVAMEY
RNAADKAKFRREHESTLILYEAAAKALKGQGVKKLPDLYALKAEYKRLAEEKERLYEKYGEAKKQMQEYG
IIKQNVDGILRMTPGKEWTQEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | N2BU79 |
Auxiliary protein
| ID | 7868 | GenBank | WP_002317278 |
| Name | WP_002317278_E8377|3 |
UniProt ID | A0A0E2H681 |
| Length | 109 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 109 a.a. Molecular weight: 12486.63 Da Isoelectric Point: 10.5610
>WP_002317278.1 MULTISPECIES: plasmid mobilization relaxosome protein MobC [Terrabacteria group]
MANRKRKIVLRVPVTPEERAMIEQKMALLHTKNFSAYARKMLIDGYIVNMDTSDIRAQTAEIQKIGVNVN
QIAKRLNGMGPVYAQDIEDIKGALAQIWQLQRYILSSQR
MANRKRKIVLRVPVTPEERAMIEQKMALLHTKNFSAYARKMLIDGYIVNMDTSDIRAQTAEIQKIGVNVN
QIAKRLNGMGPVYAQDIEDIKGALAQIWQLQRYILSSQR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A0E2H681 |
T4CP
| ID | 16491 | GenBank | WP_002317293 |
| Name | t4cp2_EQB54_RS16555_E8377|3 |
UniProt ID | _ |
| Length | 602 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 602 a.a. Molecular weight: 68060.13 Da Isoelectric Point: 6.2196
>WP_002317293.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Terrabacteria group]
MKPDVKKLLILNLPYLIFVYLFAKCGEAYRLAAGADLSGKLLHFADGFAAAFANPLPSLHLFDLCIGIAG
AVIVRLVVYCKSKNVKKYRKGMEYGSARWGTAKDIAPYIDPKFENNILLTQTERLTMTGRPKDPKTARNK
NVLVIGGSGSGKTRFFVKPNLMQCFPTTDYPTSFVVTDPKGTLVLETGRMFQQAGYRIKILNTINFSKSM
KYNPFVYIHSEKDVLKLVNTLISNTKGEGEKSAEDFWVKSERLFYSALIGYIWYEAPAEEMNFSTLLEMI
NASEAREDDPEFQSPVDLMFERLEQKDPDHFAVRQYKKFLLSAGKTRSSILISCGARLAPFDIREVRELM
EDDEMELDTIGDEKTVLYLIMSDTDTTFNFILAMLQSQLINLLCDRADDVYGGRLPVHVRLILDEFANIG
QIPNFDKLIATIRSREISASIILQSQSQLKAIYKDAAEIISDNCDSVLFLSGRGKNAKEISDALGKETID
SFNTSENRGSQTSHGLNYQKLGKELMSQDEIAVMDGGKCILQLRGVRPFLSEKFDITKHPRYKYLADADK
KNTFDVDRFLTITRRKRQQVVTQDEVFDLYEIDLSDEDAAAE
MKPDVKKLLILNLPYLIFVYLFAKCGEAYRLAAGADLSGKLLHFADGFAAAFANPLPSLHLFDLCIGIAG
AVIVRLVVYCKSKNVKKYRKGMEYGSARWGTAKDIAPYIDPKFENNILLTQTERLTMTGRPKDPKTARNK
NVLVIGGSGSGKTRFFVKPNLMQCFPTTDYPTSFVVTDPKGTLVLETGRMFQQAGYRIKILNTINFSKSM
KYNPFVYIHSEKDVLKLVNTLISNTKGEGEKSAEDFWVKSERLFYSALIGYIWYEAPAEEMNFSTLLEMI
NASEAREDDPEFQSPVDLMFERLEQKDPDHFAVRQYKKFLLSAGKTRSSILISCGARLAPFDIREVRELM
EDDEMELDTIGDEKTVLYLIMSDTDTTFNFILAMLQSQLINLLCDRADDVYGGRLPVHVRLILDEFANIG
QIPNFDKLIATIRSREISASIILQSQSQLKAIYKDAAEIISDNCDSVLFLSGRGKNAKEISDALGKETID
SFNTSENRGSQTSHGLNYQKLGKELMSQDEIAVMDGGKCILQLRGVRPFLSEKFDITKHPRYKYLADADK
KNTFDVDRFLTITRRKRQQVVTQDEVFDLYEIDLSDEDAAAE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 15430..24319
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EQB54_RS16525 | 11749..12564 | + | 816 | WP_002326780 | helix-turn-helix domain-containing protein | - |
| EQB54_RS16530 | 12690..12947 | + | 258 | WP_002326781 | hypothetical protein | - |
| EQB54_RS16535 | 13199..13675 | - | 477 | Protein_16 | recombinase family protein | - |
| EQB54_RS16540 | 13922..15121 | + | 1200 | WP_002317296 | DUF6017 domain-containing protein | - |
| EQB54_RS16545 | 15143..15355 | + | 213 | WP_002317295 | DUF5348 domain-containing protein | - |
| EQB54_RS16550 | 15430..15906 | + | 477 | WP_002317294 | PcfB family protein | cd411 |
| EQB54_RS16555 | 15903..17711 | + | 1809 | WP_002317293 | VirD4-like conjugal transfer protein, CD1115 family | - |
| EQB54_RS16560 | 17788..18003 | + | 216 | WP_002317292 | Maff2 family mobile element protein | prgF |
| EQB54_RS16565 | 18095..18958 | + | 864 | WP_002317291 | VirB6/TrbL-like conjugal transfer protein, CD1112 family | prgHa |
| EQB54_RS16570 | 18989..19531 | + | 543 | WP_002317290 | MT-A70 family methyltransferase | - |
| EQB54_RS16575 | 19545..19967 | + | 423 | WP_002370797 | PrgI family protein | prgIa |
| EQB54_RS16580 | 19897..22296 | + | 2400 | WP_002317289 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| EQB54_RS16585 | 22328..24319 | + | 1992 | WP_002382864 | C40 family peptidase | cd419a |
| EQB54_RS16590 | 24342..24593 | + | 252 | WP_002317286 | DUF4315 family protein | - |
| EQB54_RS16595 | 24583..25812 | + | 1230 | WP_002317285 | CD1107 family mobile element protein | - |
| EQB54_RS16600 | 25809..27890 | + | 2082 | WP_002317284 | DNA topoisomerase 3 | - |
Host bacterium
| ID | 22994 | GenBank | NZ_LR135403 |
| Plasmid name | E8377|3 | Incompatibility group | - |
| Plasmid size | 63798 bp | Coordinate of oriT [Strand] | 27904..27933 [-]; 35908..35935 [+] |
| Host baterium | Enterococcus faecium isolate E8377 |
Cargo genes
| Drug resistance gene | VanHBX, erm(B), aph(3')-III, ant(6)-Ia |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |