Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 122328 |
| Name | oriT1_N56454|unnamed |
| Organism | Enterococcus faecium strain N56454 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP040905 (46544..46746 [+], 203 nt) |
| oriT length | 203 nt |
| IRs (inverted repeats) | 125..130, 143..148 (AAATCC..GGATTT) 66..72, 83..89 (ACCCCCC..GGGGGGT) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 203 nt
>oriT1_N56454|unnamed
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGGCAAAAAAATAGTAGAAAATCCTTTGGTTACAAGGGATTTAGAAAATTTCGGTGTATGTCAAATGAGCTTTAAAAGTTGACATACGCCTTTTTGA
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGGCAAAAAAATAGTAGAAAATCCTTTGGTTACAAGGGATTTAGAAAATTTCGGTGTATGTCAAATGAGCTTTAAAAGTTGACATACGCCTTTTTGA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file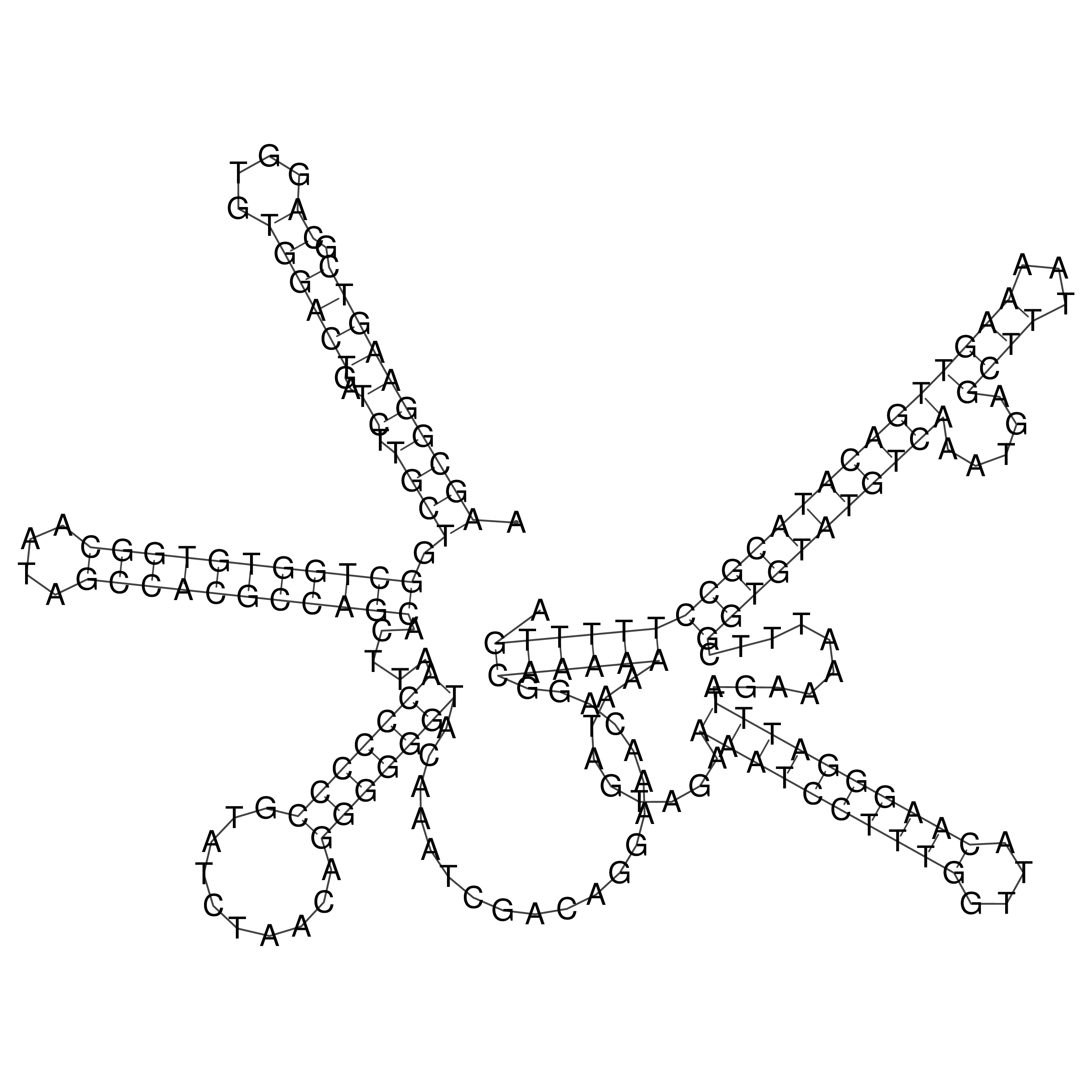
Relaxase
| ID | 14082 | GenBank | WP_139896911 |
| Name | mobT_FHK65_RS12855_N56454|unnamed |
UniProt ID | _ |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47305.96 Da Isoelectric Point: 6.7655
>WP_139896911.1 MobT family relaxase [Enterococcus faecium]
MGGISLNEQTWVQHLKEKRLSYGLSQNRLAIATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQHVVEDVLRLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MGGISLNEQTWVQHLKEKRLSYGLSQNRLAIATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQHVVEDVLRLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 22755 | GenBank | NZ_CP040905 |
| Plasmid name | N56454|unnamed | Incompatibility group | - |
| Plasmid size | 198480 bp | Coordinate of oriT [Strand] | 46544..46746 [+]; 56261..56298 [+] |
| Host baterium | Enterococcus faecium strain N56454 |
Cargo genes
| Drug resistance gene | tet(M), tet(L), erm(B), aph(3')-III, ant(6)-Ia, lnu(B), lsa(E), dfrG |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |