Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 122301 |
| Name | oriT1_pF179_2 |
| Organism | Enterococcus faecium strain F179 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP072886 (19387..19590 [+], 204 nt) |
| oriT length | 204 nt |
| IRs (inverted repeats) | 125..130, 143..148 (AAATCC..GGATTT) 66..72, 83..89 (ACCCCCC..GGGGGGT) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 204 nt
>oriT1_pF179_2
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file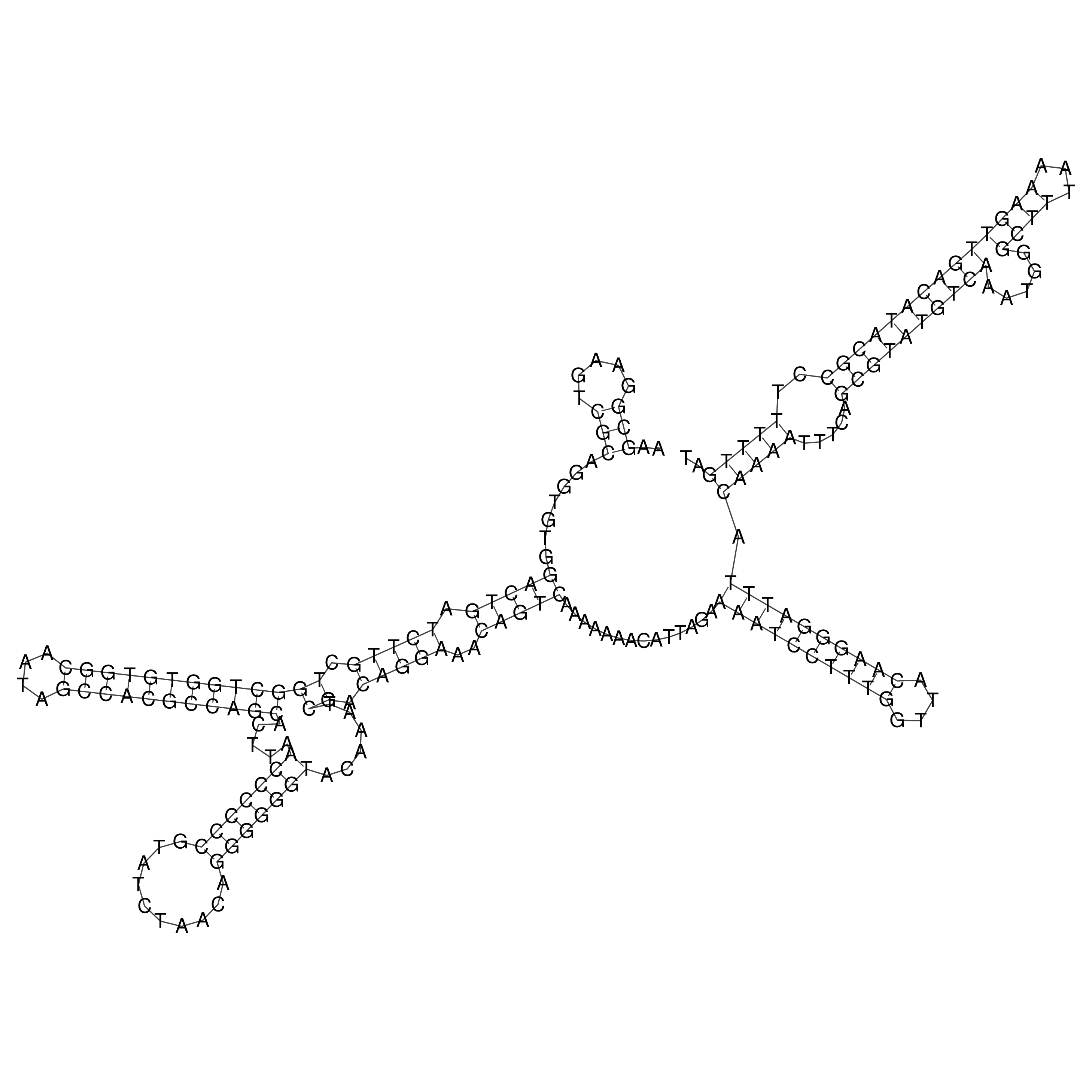
Relaxase
| ID | 14066 | GenBank | WP_000398284 |
| Name | mobT_J9540_RS13480_pF179_2 |
UniProt ID | A0A3P3PY43 |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
>WP_000398284.1 MULTISPECIES: MobT family relaxase [Bacteria]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3P3PY43 |
T4CP
| ID | 16427 | GenBank | WP_000813488 |
| Name | tcpA_J9540_RS13470_pF179_2 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>WP_000813488.1 MULTISPECIES: FtsK/SpoIIIE domain-containing protein [Bacteria]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 17282..28692
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| J9540_RS13430 (J9540_13430) | 12325..12597 | - | 273 | WP_000301765 | antitoxin | - |
| J9540_RS13435 (J9540_13435) | 12614..12829 | - | 216 | WP_001835296 | peptide-binding protein | - |
| J9540_RS13440 (J9540_13440) | 12893..13789 | - | 897 | WP_001835294 | ParA family protein | - |
| J9540_RS13445 (J9540_13445) | 13892..16036 | - | 2145 | WP_000108741 | type IA DNA topoisomerase | - |
| J9540_RS13450 (J9540_13450) | 16036..16653 | - | 618 | WP_001062589 | recombinase family protein | - |
| J9540_RS13455 (J9540_13455) | 16667..16837 | - | 171 | WP_000713594 | hypothetical protein | - |
| J9540_RS13460 (J9540_13460) | 17282..17596 | + | 315 | WP_000420682 | YdcP family protein | orf23 |
| J9540_RS13465 (J9540_13465) | 17612..17998 | + | 387 | WP_000985015 | YdcP family protein | orf23 |
| J9540_RS13470 (J9540_13470) | 18027..19412 | + | 1386 | WP_000813488 | FtsK/SpoIIIE domain-containing protein | virb4 |
| J9540_RS13475 (J9540_13475) | 19415..19567 | + | 153 | WP_000879507 | hypothetical protein | - |
| J9540_RS13480 (J9540_13480) | 19590..20795 | + | 1206 | WP_000398284 | MobT family relaxase | - |
| J9540_RS13485 (J9540_13485) | 20838..21059 | + | 222 | WP_001009056 | hypothetical protein | orf19 |
| J9540_RS13490 (J9540_13490) | 21176..21673 | + | 498 | WP_000342539 | antirestriction protein ArdA | - |
| J9540_RS13495 (J9540_13495) | 21762..22154 | + | 393 | WP_000723888 | conjugal transfer protein | orf17a |
| J9540_RS13500 (J9540_13500) | 22138..24585 | + | 2448 | WP_000331160 | ATP-binding protein | virb4 |
| J9540_RS13505 (J9540_13505) | 24588..26765 | + | 2178 | WP_001574271 | CD3337/EF1877 family mobilome membrane protein | orf15 |
| J9540_RS13510 (J9540_13510) | 26762..27763 | + | 1002 | WP_001574272 | lysozyme family protein | orf14 |
| J9540_RS13515 (J9540_13515) | 27760..28692 | + | 933 | WP_001224319 | conjugal transfer protein | orf13 |
| J9540_RS14155 | 29006..29053 | + | 48 | Protein_34 | hypothetical protein | - |
| J9540_RS13525 (J9540_13525) | 29069..30988 | + | 1920 | WP_001574275 | tetracycline resistance ribosomal protection protein Tet(M) | - |
| J9540_RS13535 (J9540_13535) | 31182..32558 | + | 1377 | WP_001574277 | tetracycline efflux MFS transporter Tet(L) | - |
| J9540_RS13540 (J9540_13540) | 33122..33376 | + | 255 | Protein_37 | plasmid recombination protein | - |
Host bacterium
| ID | 22728 | GenBank | NZ_CP072886 |
| Plasmid name | pF179_2 | Incompatibility group | - |
| Plasmid size | 53837 bp | Coordinate of oriT [Strand] | 19387..19590 [+]; 33015..33052 [+] |
| Host baterium | Enterococcus faecium strain F179 |
Cargo genes
| Drug resistance gene | lnu(B), aph(3')-III, erm(B), tet(M), tet(L), ant(6)-Ia |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |