Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 122264 |
| Name | oriT1_pJEG050 |
| Organism | Enterococcus faecium strain Efm0123 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_KR066794 (31863..31890 [-], 28 nt) |
| oriT length | 28 nt |
| IRs (inverted repeats) | 1..7, 12..18 (GGGAGTG..CACTCCC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 28 nt
>oriT1_pJEG050
GGGAGTGGCTACACTCCCTATGCTTGCT
GGGAGTGGCTACACTCCCTATGCTTGCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file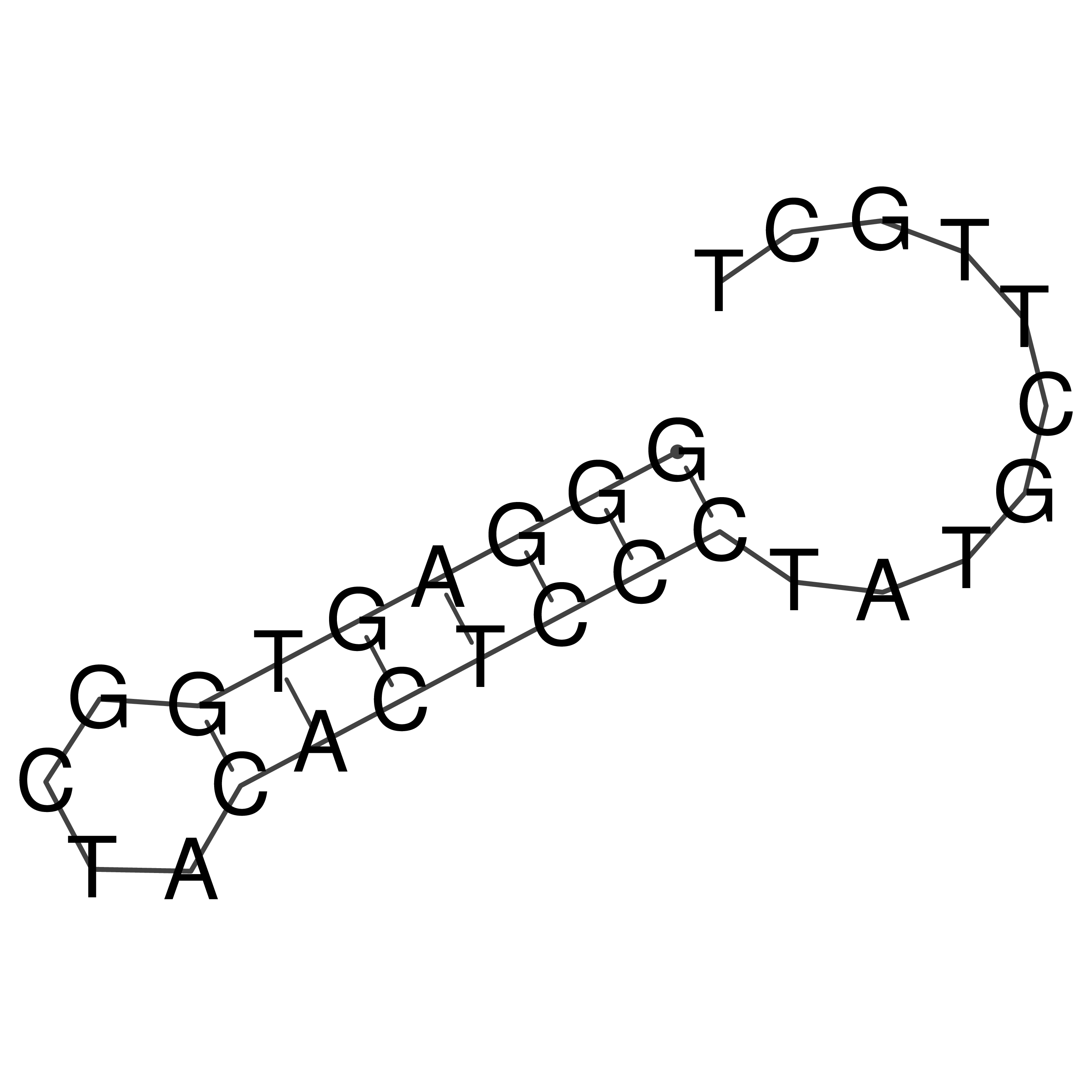
Relaxase
| ID | 14040 | GenBank | WP_002317279 |
| Name | Relaxase_HTF48_RS00230_pJEG050 |
UniProt ID | N2BU79 |
| Length | 442 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 442 a.a. Molecular weight: 50173.57 Da Isoelectric Point: 9.6754
>WP_002317279.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Terrabacteria group]
MAVTKIHPIKSTLKKALDYIENPDKTDGKLLVASFACSYETADIEFEMLLAQAYQKGNNLGHHLIQSFAP
GEATPEQAHEIGKQLAEEVLQGKYPYVLTTHIDKGHVHNHIIFCAVDMVNQRKYISNKQSYAYIRRTSDR
LCKENGLSVVKPGQSKGKSYAEWDAQRKGTSWKAKLKAAIDAAIPQAKDFDDFLRLMQAQGYEIKPGKFI
SFRAPGQERFTRCKTLGEAYTEEAIKERIKGRVITRAPKERKGISLRIDLENNIKAQQSAGYERWAKLHN
LKQAAKTLNFLTEHGIDTYPDLESKVAEITAASDEAAATLKAMEHRLADMAVLIKNISTYKQLWPVAMEY
RNAADKAKFRREHESTLILYEAAAKALKGQGVKKLPDLYALKAEYKRLAEEKERLYEKYGEAKKQMQEYG
IIKQNVDGILRMTPGKEWTQEL
MAVTKIHPIKSTLKKALDYIENPDKTDGKLLVASFACSYETADIEFEMLLAQAYQKGNNLGHHLIQSFAP
GEATPEQAHEIGKQLAEEVLQGKYPYVLTTHIDKGHVHNHIIFCAVDMVNQRKYISNKQSYAYIRRTSDR
LCKENGLSVVKPGQSKGKSYAEWDAQRKGTSWKAKLKAAIDAAIPQAKDFDDFLRLMQAQGYEIKPGKFI
SFRAPGQERFTRCKTLGEAYTEEAIKERIKGRVITRAPKERKGISLRIDLENNIKAQQSAGYERWAKLHN
LKQAAKTLNFLTEHGIDTYPDLESKVAEITAASDEAAATLKAMEHRLADMAVLIKNISTYKQLWPVAMEY
RNAADKAKFRREHESTLILYEAAAKALKGQGVKKLPDLYALKAEYKRLAEEKERLYEKYGEAKKQMQEYG
IIKQNVDGILRMTPGKEWTQEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | N2BU79 |
Auxiliary protein
| ID | 7801 | GenBank | WP_002317278 |
| Name | WP_002317278_pJEG050 |
UniProt ID | A0A0E2H681 |
| Length | 109 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 109 a.a. Molecular weight: 12486.63 Da Isoelectric Point: 10.5610
>WP_002317278.1 MULTISPECIES: plasmid mobilization relaxosome protein MobC [Terrabacteria group]
MANRKRKIVLRVPVTPEERAMIEQKMALLHTKNFSAYARKMLIDGYIVNMDTSDIRAQTAEIQKIGVNVN
QIAKRLNGMGPVYAQDIEDIKGALAQIWQLQRYILSSQR
MANRKRKIVLRVPVTPEERAMIEQKMALLHTKNFSAYARKMLIDGYIVNMDTSDIRAQTAEIQKIGVNVN
QIAKRLNGMGPVYAQDIEDIKGALAQIWQLQRYILSSQR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A0E2H681 |
Host bacterium
| ID | 22691 | GenBank | NZ_KR066794 |
| Plasmid name | pJEG050 | Incompatibility group | - |
| Plasmid size | 68175 bp | Coordinate of oriT [Strand] | 31863..31890 [-]; 39865..39894 [+] |
| Host baterium | Enterococcus faecium strain Efm0123 |
Cargo genes
| Drug resistance gene | erm(B), aph(3')-III, ant(6)-Ia, lnu(B), lsa(E), VanHBX |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |