Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 122255 |
| Name | oriT1_p11759-1 |
| Organism | Morganella morganii strain 11759 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MZ848136 (5432..6028 [-], 597 nt) |
| oriT length | 597 nt |
| IRs (inverted repeats) | 580..585, 590..595 (ATGCCC..GGGCAT) 507..513, 519..525 (CCTCCCG..CGGGAGG) 443..448, 452..457 (GTTCGC..GCGAAC) 79..84, 93..98 (GCTTTA..TAAAGC) 29..37, 46..54 (TTTTGATAA..TTATCAAAA) |
| Location of nic site | 303..304 |
| Conserved sequence flanking the nic site |
TCCTGCATCG |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 597 nt
>oriT1_p11759-1
CTTTGTTTACCTGTTAAGTACATCGTTGTTTTGATAAAATCTTTATTATCAAAAAATGTATTTATATCCTTACTGCCAGCTTTACTTTTTAATAAAGCACTTGCTAAAAGTTTCATGCCTTTCGCTACTGTTAAAGCTGGCTCATCTGGATAAAGAATTTTAAGACAATCAAGAAGTTCATCATGCTGTTGTTCTTCCAGTCTGAAATTAACTTGCTTCATAAAATCCTCCAACAAGAACCGACTGTAGGTCACCGGGCAAACGAGCGAAATGCTAGCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTGAATCTGGCACCACTCATAAATTGTGCTTGAGCATCAACAAAAAAGCAAAAAGGCTGATGTTGTCATGGCTGAAGCTACGTTCGTCTGCGACCGGAAACCCAATCAATGCGGTAAGGACAGTGCCGTCAGGTTCGCTCCGCGAACGGTCTGGCAGGAGGCGCAAGCGCAGTCCCGCGCAATATATCAGAAAGCACCTCCCGAAATACGGGAGGGCTTCGGCTATTCAGCTAAGGTTGCTCTGTTCAGGTTGCTGAGTGTAGCCTTCCATGCCCTGACGGGCATCA
CTTTGTTTACCTGTTAAGTACATCGTTGTTTTGATAAAATCTTTATTATCAAAAAATGTATTTATATCCTTACTGCCAGCTTTACTTTTTAATAAAGCACTTGCTAAAAGTTTCATGCCTTTCGCTACTGTTAAAGCTGGCTCATCTGGATAAAGAATTTTAAGACAATCAAGAAGTTCATCATGCTGTTGTTCTTCCAGTCTGAAATTAACTTGCTTCATAAAATCCTCCAACAAGAACCGACTGTAGGTCACCGGGCAAACGAGCGAAATGCTAGCATTTTGCGGCGTTTGCCCTATCCTGCATCGCAGTGAATCTGGCACCACTCATAAATTGTGCTTGAGCATCAACAAAAAAGCAAAAAGGCTGATGTTGTCATGGCTGAAGCTACGTTCGTCTGCGACCGGAAACCCAATCAATGCGGTAAGGACAGTGCCGTCAGGTTCGCTCCGCGAACGGTCTGGCAGGAGGCGCAAGCGCAGTCCCGCGCAATATATCAGAAAGCACCTCCCGAAATACGGGAGGGCTTCGGCTATTCAGCTAAGGTTGCTCTGTTCAGGTTGCTGAGTGTAGCCTTCCATGCCCTGACGGGCATCA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file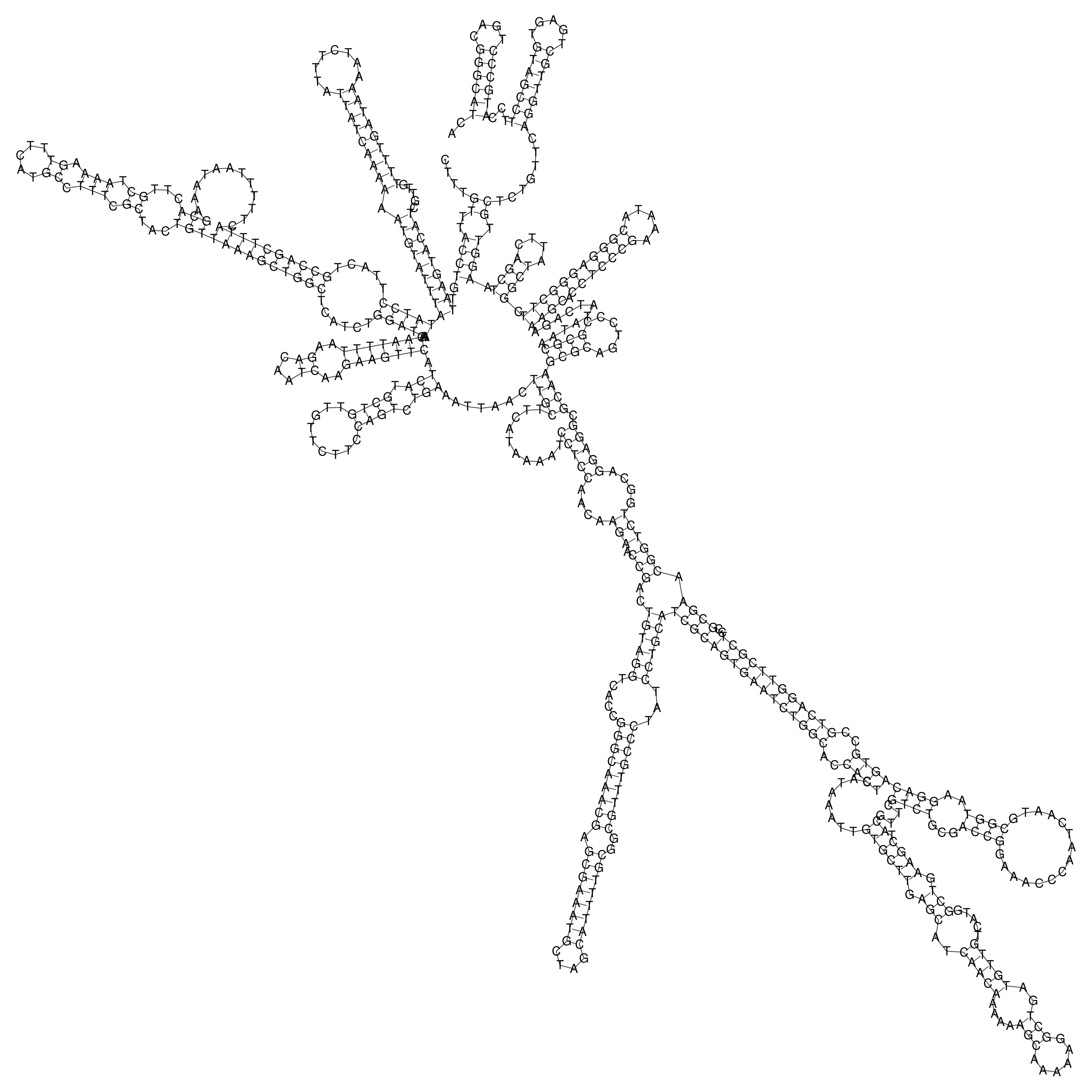
Relaxase
| ID | 14034 | GenBank | WP_000539535 |
| Name | mobP1_NLJ10_RS00070_p11759-1 |
UniProt ID | _ |
| Length | 388 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 388 a.a. Molecular weight: 44455.74 Da Isoelectric Point: 10.4644
>WP_000539535.1 MULTISPECIES: MobP1 family relaxase [Enterobacterales]
MGVYVDKEYRVKRKSSENGRKSAFAHKVKNGGKNYSRNVQERINRKGASKEVVVKISGGAITRQGVRNSI
DYMSRESELPVMSESGQVWTGDEIQEAKEHMIDRANDPQHVINDKGKENKKITQNIVFSPPVSAKVKPED
LLESVRKTMQKKYPNHRFVLGYHCDKKEHPHVHVVFRIRDNDGKRADIRKKDLREIRTGFCEELKLRGYD
VKATHKQQHGLNQSVKDAHNTAPKRQKGVYEVVDVGYDHYQNDKTKSKQYFIKLKTLNKGVEKTYWGADF
GDLCSRESVKAGDLVRLKKLGQKEVKIPALDKNGVQHGWKTVHRNEWQLENLGVKGIDRTPSASKELVLN
SPDMLLKQQQRMAQFTQQKASTLQSEQKLKTGIKFLGL
MGVYVDKEYRVKRKSSENGRKSAFAHKVKNGGKNYSRNVQERINRKGASKEVVVKISGGAITRQGVRNSI
DYMSRESELPVMSESGQVWTGDEIQEAKEHMIDRANDPQHVINDKGKENKKITQNIVFSPPVSAKVKPED
LLESVRKTMQKKYPNHRFVLGYHCDKKEHPHVHVVFRIRDNDGKRADIRKKDLREIRTGFCEELKLRGYD
VKATHKQQHGLNQSVKDAHNTAPKRQKGVYEVVDVGYDHYQNDKTKSKQYFIKLKTLNKGVEKTYWGADF
GDLCSRESVKAGDLVRLKKLGQKEVKIPALDKNGVQHGWKTVHRNEWQLENLGVKGIDRTPSASKELVLN
SPDMLLKQQQRMAQFTQQKASTLQSEQKLKTGIKFLGL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16421 | GenBank | WP_000053829 |
| Name | t4cp2_NLJ10_RS00135_p11759-1 |
UniProt ID | _ |
| Length | 611 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 611 a.a. Molecular weight: 69290.20 Da Isoelectric Point: 6.9121
>WP_000053829.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Enterobacterales]
MSLKLPDKGQWVFIGLVMCLVTYYAGSVAVYFLNGKTPLYIWKNFDSMLLWKIITESNIRTDIRLTAIPS
LLSGMVSSLIVPVFVIWQLNKMDVALYGDAKFASDNDLKKSKLLKWETENDTDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREILLGNKVYLLDPFNSKTHQFNPLF
YIDLKAESGAKDLLKLIEILFPSYGMTGAEAHFNNLAGQYWTGLAKLLHFFINYEPSWLNEFGLKPVFSI
GSVVDLYSNIDRELILSKREELEGTNGLDENALYHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDITVYVGVNAEDISLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGIEGAKTLMSAHPCRIIYAVSEEDDAAKIS
EKLGYITTTSKSTSKSRGRSASQGESESEARRALVLPQELGTLDFKEEFIILKGENPVKAEKALYYLDPY
FMDRLMKVSPKLASLTMELNKTKKIFGVKGLKYPSKEKMLSVGELESEVLL
MSLKLPDKGQWVFIGLVMCLVTYYAGSVAVYFLNGKTPLYIWKNFDSMLLWKIITESNIRTDIRLTAIPS
LLSGMVSSLIVPVFVIWQLNKMDVALYGDAKFASDNDLKKSKLLKWETENDTDILVGAYKGKYLWYTAPD
FVSLGAGTRAGKGAAIGIPNLLVRKHSLIALDPKQELWKITSKVREILLGNKVYLLDPFNSKTHQFNPLF
YIDLKAESGAKDLLKLIEILFPSYGMTGAEAHFNNLAGQYWTGLAKLLHFFINYEPSWLNEFGLKPVFSI
GSVVDLYSNIDRELILSKREELEGTNGLDENALYHLRDALTKIREYHETEDEQRSSIDGSFRKKMSLFYL
PTVRKCTDGNDFDLRQLRREDITVYVGVNAEDISLAYDFLNLFFNFVVEVTLRENPDFDPTLKHDCLMFL
DEFPSIGYMPIIKKGSGYIAGFKLKLLTIYQNISQLNEIYGIEGAKTLMSAHPCRIIYAVSEEDDAAKIS
EKLGYITTTSKSTSKSRGRSASQGESESEARRALVLPQELGTLDFKEEFIILKGENPVKAEKALYYLDPY
FMDRLMKVSPKLASLTMELNKTKKIFGVKGLKYPSKEKMLSVGELESEVLL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 8585..18545
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NLJ10_RS00035 | 3601..3954 | + | 354 | WP_024193965 | hypothetical protein | - |
| NLJ10_RS00040 | 4037..4255 | + | 219 | WP_001407769 | hypothetical protein | - |
| NLJ10_RS00045 | 4265..4681 | + | 417 | WP_012421208 | tail-associated lysozyme | - |
| NLJ10_RS00050 | 4696..4989 | + | 294 | WP_021568250 | hypothetical protein | - |
| NLJ10_RS00055 | 5010..5207 | + | 198 | WP_012599846 | hypothetical protein | - |
| NLJ10_RS00060 | 5220..5492 | + | 273 | WP_001547192 | hypothetical protein | - |
| NLJ10_RS00065 | 5808..6353 | + | 546 | WP_000815825 | DNA distortion polypeptide 1 | - |
| NLJ10_RS00070 | 6356..7522 | + | 1167 | WP_000539535 | MobP1 family relaxase | - |
| NLJ10_RS00075 | 7855..8361 | + | 507 | WP_000872474 | transcription termination/antitermination NusG family protein | - |
| NLJ10_RS00080 | 8585..9232 | + | 648 | WP_000953535 | lytic transglycosylase domain-containing protein | virB1 |
| NLJ10_RS00085 | 9210..9506 | + | 297 | WP_075332605 | TrbC/VirB2 family protein | virB2 |
| NLJ10_RS00090 | 9525..12278 | + | 2754 | WP_001363160 | VirB3 family type IV secretion system protein | virb4 |
| NLJ10_RS00095 | 12289..13047 | + | 759 | WP_000744201 | type IV secretion system protein | - |
| NLJ10_RS00100 | 13048..13275 | + | 228 | WP_000835348 | EexN family lipoprotein | - |
| NLJ10_RS00105 | 13285..14418 | + | 1134 | WP_001547182 | type IV secretion system protein | virB6 |
| NLJ10_RS00115 | 14663..15376 | + | 714 | WP_000394613 | type IV secretion system protein | virB8 |
| NLJ10_RS00120 | 15381..16310 | + | 930 | WP_000783381 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| NLJ10_RS00125 | 16307..17512 | + | 1206 | WP_001547183 | VirB10/TraB/TrbI family type IV secretion system protein | virB10 |
| NLJ10_RS00130 | 17514..18545 | + | 1032 | WP_001058476 | P-type DNA transfer ATPase VirB11 | virB11 |
| NLJ10_RS00135 | 18548..20383 | + | 1836 | WP_000053829 | type IV secretory system conjugative DNA transfer family protein | - |
| NLJ10_RS00140 | 20380..20793 | + | 414 | WP_001547184 | cag pathogenicity island Cag12 family protein | - |
| NLJ10_RS00145 | 20790..21092 | + | 303 | WP_000717623 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| NLJ10_RS00150 | 21098..21355 | + | 258 | WP_001547185 | hypothetical protein | - |
| NLJ10_RS00155 | 21458..21766 | + | 309 | WP_000647576 | hypothetical protein | - |
| NLJ10_RS00160 | 21763..22257 | + | 495 | WP_001215682 | thermonuclease family protein | - |
| NLJ10_RS00165 | 22273..22677 | + | 405 | WP_001218539 | hypothetical protein | - |
| NLJ10_RS00170 | 22737..23057 | + | 321 | WP_001020837 | hypothetical protein | - |
Host bacterium
| ID | 22682 | GenBank | NZ_MZ848136 |
| Plasmid name | p11759-1 | Incompatibility group | IncX1 |
| Plasmid size | 33731 bp | Coordinate of oriT [Strand] | 5432..6028 [-]; 31357..31779 [-] |
| Host baterium | Morganella morganii strain 11759 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |