Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 122182 |
| Name | oriT_51271924|unnamed |
| Organism | Enterococcus faecalis strain 51271924 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JANHGF010000006 (34149..34198 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | 34..35 |
| Conserved sequence flanking the nic site |
GCTTGCAAAA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_51271924|unnamed
AATATCGCAACATGGTACCATGTTGCTCCGCTTGCAAAAAGAAAGCCTAC
AATATCGCAACATGGTACCATGTTGCTCCGCTTGCAAAAAGAAAGCCTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file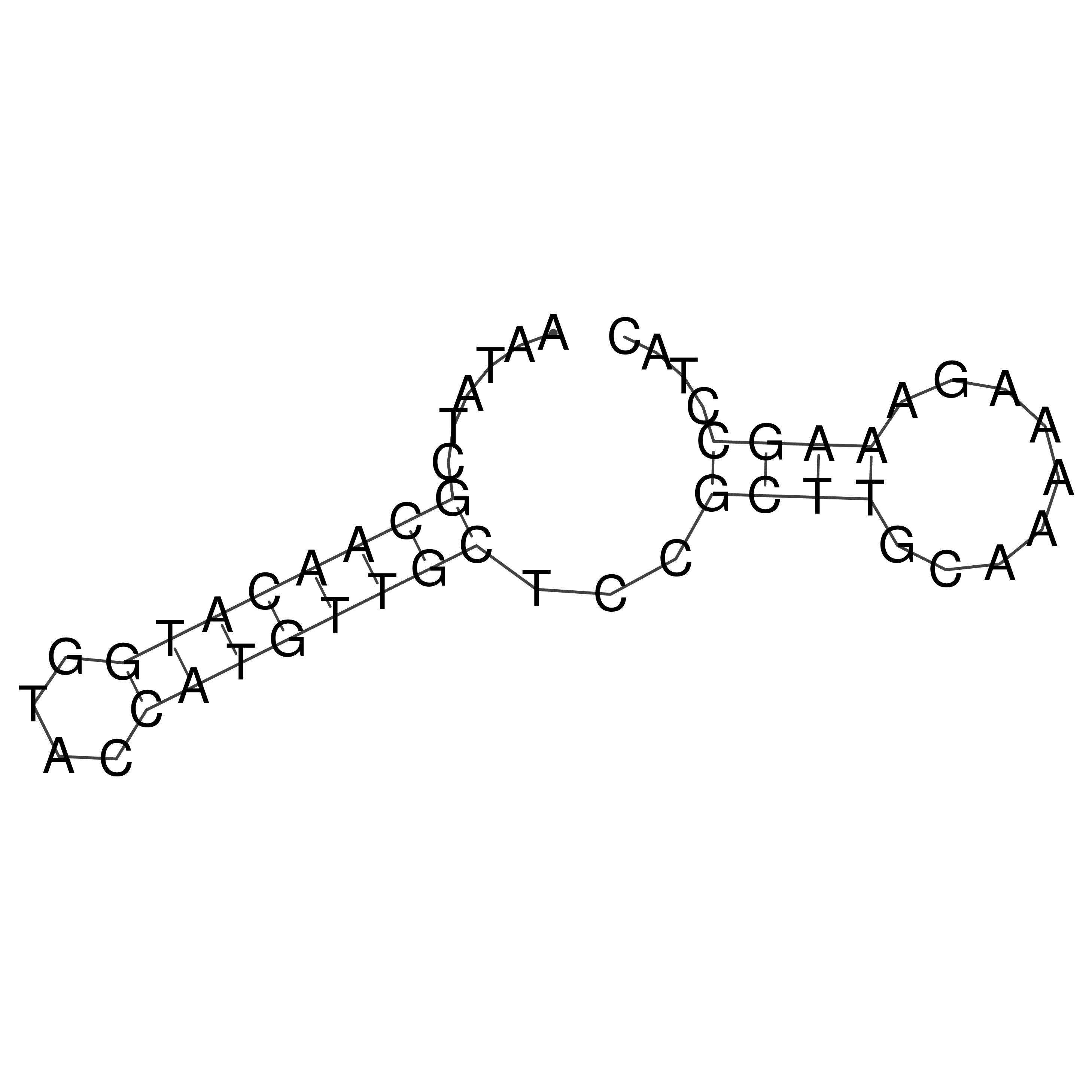
Relaxase
| ID | 13979 | GenBank | WP_010774278 |
| Name | Relaxase_NPQ23_RS17270_51271924|unnamed |
UniProt ID | Q82YL3 |
| Length | 561 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 561 a.a. Molecular weight: 65308.93 Da Isoelectric Point: 6.1201
>WP_010774278.1 relaxase/mobilization nuclease domain-containing protein [Enterococcus faecalis]
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKKVFEQISDKIAAKEGAKIIEKSPKQT
HTKYTKWQTESIYKQKIKSRLDFLLEQSSSINDFLEKAEALNLSADFSNKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIIERLKENKNVLTVEECVKRYEEKNEQEKNNFDYQFTIEPWQLSHKTERGYYINIDYG
YGNSGKLFVGGYKVDPLANGNYNIYIKRNEYFYFMSDKDATKSKYLTGASLIKQLRLYNGQTPLKREPVM
QTIDELVSAINFLAANEIEDTRQLKLLEEKLEVAFLEAEKTLETLDEKMLELHQLSNLLLENELQGDPEL
IQKKLKTLLPDATLAEFSYEDVRGEIEAIKTSQTLLESKLERTRNEINQLHEIQAVQEKEPEKQTDIKPK
L
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKKVFEQISDKIAAKEGAKIIEKSPKQT
HTKYTKWQTESIYKQKIKSRLDFLLEQSSSINDFLEKAEALNLSADFSNKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIIERLKENKNVLTVEECVKRYEEKNEQEKNNFDYQFTIEPWQLSHKTERGYYINIDYG
YGNSGKLFVGGYKVDPLANGNYNIYIKRNEYFYFMSDKDATKSKYLTGASLIKQLRLYNGQTPLKREPVM
QTIDELVSAINFLAANEIEDTRQLKLLEEKLEVAFLEAEKTLETLDEKMLELHQLSNLLLENELQGDPEL
IQKKLKTLLPDATLAEFSYEDVRGEIEAIKTSQTLLESKLERTRNEINQLHEIQAVQEKEPEKQTDIKPK
L
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q82YL3 |
Auxiliary protein
| ID | 7778 | GenBank | WP_002367696 |
| Name | WP_002367696_51271924|unnamed |
UniProt ID | Q82YL4 |
| Length | 118 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 118 a.a. Molecular weight: 14042.10 Da Isoelectric Point: 9.9724
>WP_002367696.1 MULTISPECIES: plasmid mobilization relaxosome protein MobC [Bacteria]
MENKQKLKRPIQRKIRFSKEENELINRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKEQS
MENKQKLKRPIQRKIRFSKEENELINRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKEQS
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q82YL4 |
T4CP
| ID | 16405 | GenBank | WP_002425085 |
| Name | t4cp2_NPQ23_RS17250_51271924|unnamed |
UniProt ID | _ |
| Length | 609 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 609 a.a. Molecular weight: 70145.14 Da Isoelectric Point: 6.1811
>WP_002425085.1 VirD4-like conjugal transfer protein, CD1115 family [Enterococcus faecalis]
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEGEKTTGEDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRVLEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEGEKTTGEDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRVLEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 14217..31505
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NPQ23_RS17155 (NPQ23_17105) | 10079..10459 | + | 381 | WP_002365885 | PrgR | - |
| NPQ23_RS17160 (NPQ23_17110) | 10510..10731 | + | 222 | WP_016616022 | hypothetical protein | - |
| NPQ23_RS17165 (NPQ23_17115) | 11033..11338 | + | 306 | Protein_13 | hypothetical protein | - |
| NPQ23_RS17170 (NPQ23_17120) | 11349..14024 | + | 2676 | WP_011109619 | surface exclusion protein SEA1/PrgA | - |
| NPQ23_RS17175 (NPQ23_17125) | 14217..18134 | + | 3918 | WP_002395058 | LPXTG-anchored aggregation substance PrgB/Asc10 | prgB |
| NPQ23_RS17180 (NPQ23_17130) | 18196..18552 | + | 357 | WP_002367672 | pheromone response system RNA-binding regulator PrgU | - |
| NPQ23_RS17185 (NPQ23_17135) | 18580..19437 | + | 858 | WP_002367673 | pheromone response LPXTG-anchored adhesin PrgC | prgC |
| NPQ23_RS17190 (NPQ23_17140) | 19479..20378 | + | 900 | WP_011109620 | hypothetical protein | - |
| NPQ23_RS17195 (NPQ23_17145) | 20398..20832 | + | 435 | WP_002365857 | hypothetical protein | - |
| NPQ23_RS17200 (NPQ23_17150) | 20879..21106 | + | 228 | WP_002365858 | hypothetical protein | - |
| NPQ23_RS17205 (NPQ23_17155) | 21120..21416 | + | 297 | WP_002387740 | hypothetical protein | - |
| NPQ23_RS17210 (NPQ23_17160) | 21431..22231 | + | 801 | WP_010774271 | type IV secretion system protein | prgHb |
| NPQ23_RS17215 (NPQ23_17165) | 22233..22586 | + | 354 | WP_010774272 | PrgI family protein | prgIc |
| NPQ23_RS17220 (NPQ23_17170) | 22540..24930 | + | 2391 | WP_010774273 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| NPQ23_RS17225 (NPQ23_17175) | 24942..27557 | + | 2616 | WP_010774274 | phage tail tip lysozyme | prgK |
| NPQ23_RS17230 (NPQ23_17180) | 27581..28207 | + | 627 | WP_002395055 | hypothetical protein | prgL |
| NPQ23_RS17235 (NPQ23_17185) | 28194..28439 | + | 246 | WP_002365866 | hypothetical protein | - |
| NPQ23_RS17240 (NPQ23_17190) | 28423..29031 | + | 609 | WP_010774275 | hypothetical protein | - |
| NPQ23_RS17245 (NPQ23_17195) | 29191..29676 | + | 486 | WP_010774276 | DUF3801 domain-containing protein | gbs1365 |
| NPQ23_RS17250 (NPQ23_17200) | 29676..31505 | + | 1830 | WP_002425085 | VirD4-like conjugal transfer protein, CD1115 family | virb4 |
| NPQ23_RS17255 (NPQ23_17205) | 31555..33714 | + | 2160 | WP_010774277 | ArdC-like ssDNA-binding domain-containing protein | - |
| NPQ23_RS17260 (NPQ23_17210) | 33747..34019 | + | 273 | WP_002362409 | LtrD | - |
| NPQ23_RS17265 (NPQ23_17215) | 34265..34621 | + | 357 | WP_002367696 | plasmid mobilization relaxosome protein MobC | - |
| NPQ23_RS17270 (NPQ23_17220) | 34622..36307 | + | 1686 | WP_010774278 | relaxase/mobilization nuclease domain-containing protein | - |
Host bacterium
| ID | 22609 | GenBank | NZ_JANHGF010000006 |
| Plasmid name | 51271924|unnamed | Incompatibility group | - |
| Plasmid size | 57661 bp | Coordinate of oriT [Strand] | 34149..34198 [+] |
| Host baterium | Enterococcus faecalis strain 51271924 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | prgB/asc10 |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21, AcrIIA9 |