Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121985 |
| Name | oriT_pKPYL1 |
| Organism | Raoultella ornithinolytica strain Yangling I2 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013339 (77329..77378 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pKPYL1
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file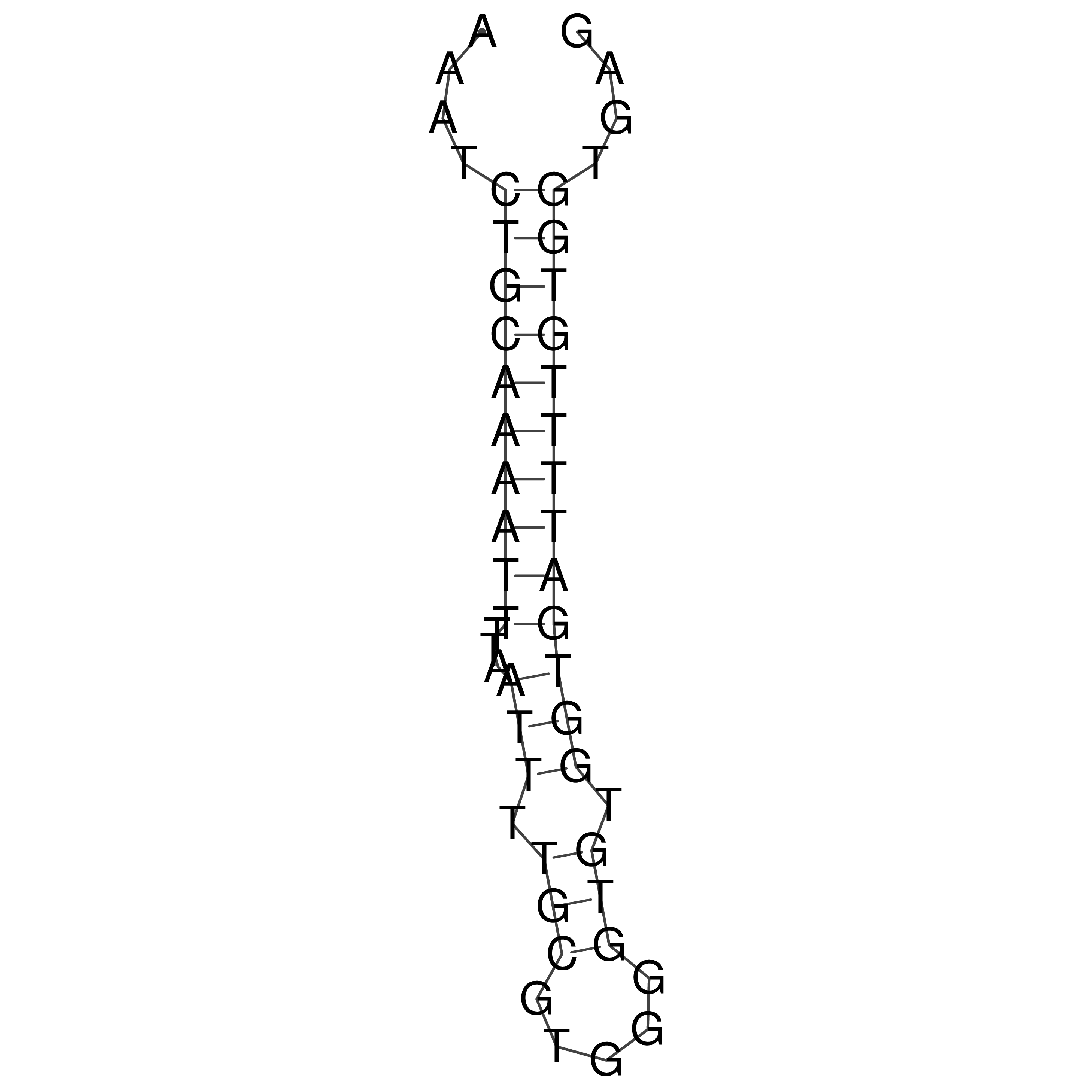
Relaxase
| ID | 13860 | GenBank | WP_064357876 |
| Name | traI_ATN83_RS26015_pKPYL1 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190310.03 Da Isoelectric Point: 6.1562
>WP_064357876.1 conjugative transfer relaxase/helicase TraI [Raoultella ornithinolytica]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKPVDKAVFTEILKGKLPDGSDLTRM
QDGTNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVTKALQQVETLAATRVMTDGKSETVLTGNL
IVAKFTHDTNRNEEPQLHTHAVVMNATQNGDKWQSLGTDTVGKTGFIENVYANQIAFGKIYREELKPLVE
ILGYKTEVVGKHGMWEMNEVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKAAIDPAEKMVEWMN
TMKETGFDMRSYREDADKRAAELASAPVSPVKTDGPDISDVVTKAIASLSDRKVQFTYADLLARTVSQLE
AKPGVFEQARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVQRQNHVSVTPDASVVRQAP
FSDAVSVLAQDRPAMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDARLAGEAVT
GKSALQDGTAFIPGGTLIIDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQVTADIISEPDKGARYSLLAQEFAASVREGQESVAQISGTREQNVLNSVIRNTLKNEGVLGEKE
MTVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFVIDRVTASSNMLTLKDKEGERLDMKVSAV
DSQWTLFRADKLPVAEGERLAVLGKIPDTRLKGGESITVLKSGDGQLTVQRPGQKTTQSLSVGSSPFDGI
KVGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTMEKLARHTAFSVVSE
QLKARSGETDLDAAIAQQKAGLHTPAEQAIHLSVPLLESDDLTFTRPQLLATALETGGGKVSMADIDTTI
QAQIRSGQLLNVSVSSGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTGLTAGQRASTR
MILETPDRFTVVQGYAGVGKTTQFRAVTSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLLRNGQTPDFSNTLFLIDESSMVGLSDKAKALSLIAAGGGRAVSSGDTDQLQSISPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIDRDVHRALTTIEQVTPEQVLRKEGAWAPGSSVVEFTRAQEKAIQ
EALKKGESVPAGQPATLYEALVKDYAGRTPEAQSQTLVITHLNKDRRALNGLIHDARRENGETGKEEITL
PVLVTSNIRDGELRRLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRREDITVSEGDRMRFSKSDPERGYVANSVWEVKSVSGDSVTLSDGKTSRTLNPKADEAQRHIDLAYAIT
AHGAQGASEPFAIALEGVAEGRKAMASFESAYVGLSRMKQHVQVYTDSREGWIKAINASPSKATAHDILA
PRNDRAVHTAEQLFSRARPMNETAAGRAALQQSGLAKGISPGKFISPGKQYPQPHVALPVYDKNGKEAGI
WLSSLTDSDGRLQVMGGEGRVMGNEDARFVALQGSRNGESLQAGNMGEGVRVARDNPDSGVVVCLAGDDR
PWNPGAITGGRVWADPIRVVSPEAAGTDIPLPPEVLAQRAAEEAQRQNMEKQAEQTAREVAGDGRKAGES
EDRVKEVIGDVIRGLEREKPGEEKMTLPDDPQTRKQEAAVQQVAQESLQRERLQQMERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKPVDKAVFTEILKGKLPDGSDLTRM
QDGTNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVTKALQQVETLAATRVMTDGKSETVLTGNL
IVAKFTHDTNRNEEPQLHTHAVVMNATQNGDKWQSLGTDTVGKTGFIENVYANQIAFGKIYREELKPLVE
ILGYKTEVVGKHGMWEMNEVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKAAIDPAEKMVEWMN
TMKETGFDMRSYREDADKRAAELASAPVSPVKTDGPDISDVVTKAIASLSDRKVQFTYADLLARTVSQLE
AKPGVFEQARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVQRQNHVSVTPDASVVRQAP
FSDAVSVLAQDRPAMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDARLAGEAVT
GKSALQDGTAFIPGGTLIIDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQVTADIISEPDKGARYSLLAQEFAASVREGQESVAQISGTREQNVLNSVIRNTLKNEGVLGEKE
MTVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFVIDRVTASSNMLTLKDKEGERLDMKVSAV
DSQWTLFRADKLPVAEGERLAVLGKIPDTRLKGGESITVLKSGDGQLTVQRPGQKTTQSLSVGSSPFDGI
KVGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTMEKLARHTAFSVVSE
QLKARSGETDLDAAIAQQKAGLHTPAEQAIHLSVPLLESDDLTFTRPQLLATALETGGGKVSMADIDTTI
QAQIRSGQLLNVSVSSGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTGLTAGQRASTR
MILETPDRFTVVQGYAGVGKTTQFRAVTSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLLRNGQTPDFSNTLFLIDESSMVGLSDKAKALSLIAAGGGRAVSSGDTDQLQSISPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIDRDVHRALTTIEQVTPEQVLRKEGAWAPGSSVVEFTRAQEKAIQ
EALKKGESVPAGQPATLYEALVKDYAGRTPEAQSQTLVITHLNKDRRALNGLIHDARRENGETGKEEITL
PVLVTSNIRDGELRRLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRREDITVSEGDRMRFSKSDPERGYVANSVWEVKSVSGDSVTLSDGKTSRTLNPKADEAQRHIDLAYAIT
AHGAQGASEPFAIALEGVAEGRKAMASFESAYVGLSRMKQHVQVYTDSREGWIKAINASPSKATAHDILA
PRNDRAVHTAEQLFSRARPMNETAAGRAALQQSGLAKGISPGKFISPGKQYPQPHVALPVYDKNGKEAGI
WLSSLTDSDGRLQVMGGEGRVMGNEDARFVALQGSRNGESLQAGNMGEGVRVARDNPDSGVVVCLAGDDR
PWNPGAITGGRVWADPIRVVSPEAAGTDIPLPPEVLAQRAAEEAQRQNMEKQAEQTAREVAGDGRKAGES
EDRVKEVIGDVIRGLEREKPGEEKMTLPDDPQTRKQEAAVQQVAQESLQRERLQQMERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 7686 | GenBank | WP_064357868 |
| Name | WP_064357868_pKPYL1 |
UniProt ID | _ |
| Length | 129 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 129 a.a. Molecular weight: 14633.57 Da Isoelectric Point: 4.5352
>WP_064357868.1 conjugal transfer relaxosome DNA-binding protein TraM [Raoultella ornithinolytica]
MARVQAYASDEVAEKINAIVEKRKAEGAKDKDVSFSSVASMLLELGLRVYEAQMERKESGFNQAEFNKML
LENVIKTQFSISKVLAMESLSPHIQGDERFDFKSMVMNIRDDAKEVVERFFPDTDDEGS
MARVQAYASDEVAEKINAIVEKRKAEGAKDKDVSFSSVASMLLELGLRVYEAQMERKESGFNQAEFNKML
LENVIKTQFSISKVLAMESLSPHIQGDERFDFKSMVMNIRDDAKEVVERFFPDTDDEGS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16228 | GenBank | WP_032716692 |
| Name | traD_ATN83_RS26010_pKPYL1 |
UniProt ID | _ |
| Length | 776 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 776 a.a. Molecular weight: 86758.82 Da Isoelectric Point: 4.9734
>WP_032716692.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Klebsiella/Raoultella group]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFIIFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTINYYGRSLEYNAEQILADKYTIWCGEQLWTSFVLAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLSLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRSLDTRVDTRLSALLEAREAEGSLARALFTPDAPAPDPAEKDVKVGNTPEQSQPAE
SIESPATVKVPATAKTPAADEIGQSPETPEPAAPVTKVVTVPLIRPRSATATGTAAVATTAASTTSATTV
PAGGTEQELAQQPTEQGQDRLPAGMNDDGVIEDMDAYDAWLAGEQTQRDMQRREEVNINHSHRRDEQDDI
EIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFIIFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTINYYGRSLEYNAEQILADKYTIWCGEQLWTSFVLAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLSLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRSLDTRVDTRLSALLEAREAEGSLARALFTPDAPAPDPAEKDVKVGNTPEQSQPAE
SIESPATVKVPATAKTPAADEIGQSPETPEPAAPVTKVVTVPLIRPRSATATGTAAVATTAASTTSATTV
PAGGTEQELAQQPTEQGQDRLPAGMNDDGVIEDMDAYDAWLAGEQTQRDMQRREEVNINHSHRRDEQDDI
EIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 76776..107123
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ATN83_RS25805 (ATN83_p10099) | 71878..72606 | + | 729 | WP_064357862 | plasmid SOS inhibition protein A | - |
| ATN83_RS28970 | 72603..72762 | + | 160 | Protein_76 | theronine dehydrogenase | - |
| ATN83_RS25810 (ATN83_p10100) | 73010..73339 | + | 330 | WP_004118963 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| ATN83_RS25815 (ATN83_p10101) | 73320..73601 | + | 282 | WP_004118961 | helix-turn-helix transcriptional regulator | - |
| ATN83_RS25820 (ATN83_p10104) | 74420..74695 | + | 276 | WP_032700867 | hypothetical protein | - |
| ATN83_RS25825 (ATN83_p10105) | 74752..74910 | + | 159 | WP_162180130 | hypothetical protein | - |
| ATN83_RS25830 (ATN83_p10106) | 74936..75283 | + | 348 | WP_032700835 | hypothetical protein | - |
| ATN83_RS25835 (ATN83_p10107) | 75377..75523 | + | 147 | WP_064357864 | hypothetical protein | - |
| ATN83_RS25840 (ATN83_p10110) | 76208..76738 | + | 531 | WP_064357865 | antirestriction protein | - |
| ATN83_RS25845 (ATN83_p10111) | 76776..77255 | - | 480 | WP_032716648 | transglycosylase SLT domain-containing protein | virB1 |
| ATN83_RS25850 (ATN83_p10112) | 77680..78069 | + | 390 | WP_064357868 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| ATN83_RS28040 (ATN83_p10113) | 78270..78959 | + | 690 | WP_071887219 | hypothetical protein | - |
| ATN83_RS28045 (ATN83_p10114) | 79138..79293 | + | 156 | WP_227502495 | TraY domain-containing protein | - |
| ATN83_RS25855 (ATN83_p10115) | 79366..79734 | + | 369 | WP_032716651 | type IV conjugative transfer system pilin TraA | - |
| ATN83_RS25860 (ATN83_p10116) | 79748..80053 | + | 306 | WP_020323523 | type IV conjugative transfer system protein TraL | traL |
| ATN83_RS25865 (ATN83_p10117) | 80073..80639 | + | 567 | WP_032700829 | type IV conjugative transfer system protein TraE | traE |
| ATN83_RS25870 (ATN83_p10118) | 80626..81360 | + | 735 | WP_032700828 | type-F conjugative transfer system secretin TraK | traK |
| ATN83_RS25875 (ATN83_p10119) | 81360..82778 | + | 1419 | WP_032716653 | F-type conjugal transfer pilus assembly protein TraB | traB |
| ATN83_RS28630 (ATN83_p10120) | 82771..83553 | + | 783 | WP_155522187 | conjugal transfer protein TraP | - |
| ATN83_RS28050 (ATN83_p10121) | 83572..84141 | + | 570 | WP_071887221 | type IV conjugative transfer system lipoprotein TraV | traV |
| ATN83_RS28635 (ATN83_p10123) | 84503..84787 | + | 285 | WP_128910320 | hypothetical protein | - |
| ATN83_RS28975 (ATN83_p10124) | 84821..85300 | + | 480 | WP_227502492 | hypothetical protein | - |
| ATN83_RS25895 (ATN83_p10126) | 85781..86002 | + | 222 | WP_032716655 | hypothetical protein | - |
| ATN83_RS25905 (ATN83_p10128) | 86115..87235 | + | 1121 | WP_152292706 | IS3-like element ISKpn34 family transposase | - |
| ATN83_RS28980 (ATN83_p10129) | 87239..87628 | + | 390 | WP_064357870 | hypothetical protein | - |
| ATN83_RS25915 (ATN83_p10130) | 87641..87961 | + | 321 | WP_227502491 | hypothetical protein | - |
| ATN83_RS25920 (ATN83_p10132) | 88442..91081 | + | 2640 | WP_064357871 | type IV secretion system protein TraC | virb4 |
| ATN83_RS25925 (ATN83_p10133) | 91081..91473 | + | 393 | WP_032716659 | type-F conjugative transfer system protein TrbI | - |
| ATN83_RS25930 (ATN83_p10134) | 91470..92099 | + | 630 | WP_032716660 | type-F conjugative transfer system protein TraW | traW |
| ATN83_RS25935 (ATN83_p10135) | 92140..93099 | + | 960 | WP_032716690 | conjugal transfer pilus assembly protein TraU | traU |
| ATN83_RS25940 (ATN83_p10136) | 93112..93741 | + | 630 | WP_032700817 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| ATN83_RS25945 (ATN83_p10137) | 93741..95696 | + | 1956 | WP_032716661 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| ATN83_RS25950 (ATN83_p10138) | 95712..96092 | + | 381 | WP_032700815 | hypothetical protein | - |
| ATN83_RS25955 (ATN83_p10139) | 96082..96318 | + | 237 | WP_032700814 | conjugal transfer protein TrbE | - |
| ATN83_RS25965 (ATN83_p10140) | 96544..96870 | + | 327 | WP_032700812 | hypothetical protein | - |
| ATN83_RS25970 (ATN83_p10141) | 96893..97645 | + | 753 | WP_032724578 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| ATN83_RS25975 (ATN83_p10142) | 97656..97892 | + | 237 | WP_032700810 | type-F conjugative transfer system pilin chaperone TraQ | - |
| ATN83_RS25980 (ATN83_p10143) | 97867..98433 | + | 567 | WP_032716665 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| ATN83_RS25985 (ATN83_p10144) | 98426..98854 | + | 429 | WP_032716666 | conjugal transfer protein TrbF | - |
| ATN83_RS25990 (ATN83_p10145) | 98841..100211 | + | 1371 | WP_064357873 | conjugal transfer pilus assembly protein TraH | traH |
| ATN83_RS25995 (ATN83_p10146) | 100211..103033 | + | 2823 | WP_064357874 | conjugal transfer mating-pair stabilization protein TraG | traG |
| ATN83_RS26000 (ATN83_p10147) | 103059..103673 | + | 615 | WP_032716668 | hypothetical protein | - |
| ATN83_RS26005 (ATN83_p10148) | 103868..104599 | + | 732 | WP_032716670 | conjugal transfer complement resistance protein TraT | - |
| ATN83_RS26010 (ATN83_p10149) | 104793..107123 | + | 2331 | WP_032716692 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 22412 | GenBank | NZ_CP013339 |
| Plasmid name | pKPYL1 | Incompatibility group | IncFIB |
| Plasmid size | 152010 bp | Coordinate of oriT [Strand] | 77329..77378 [-] |
| Host baterium | Raoultella ornithinolytica strain Yangling I2 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsC, arsB, arsA, arsD, arsR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |