Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121979 |
| Name | oriT_pAtS4c |
| Organism | Allorhizobium ampelinum S4 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_011984 (145285..145324 [+], 40 nt) |
| oriT length | 40 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 40 nt
>oriT_pAtS4c
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCTTGC
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file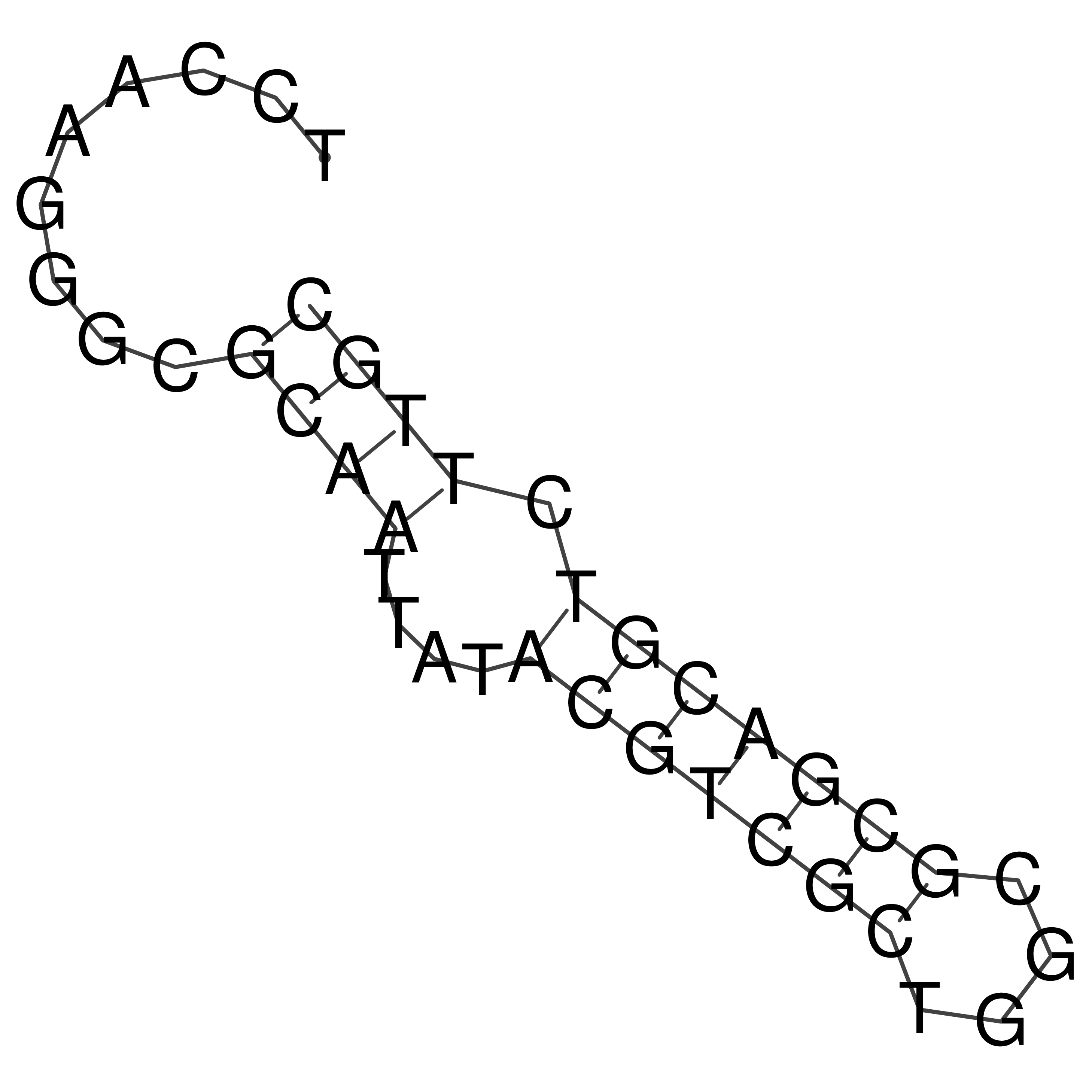
Relaxase
| ID | 13853 | GenBank | WP_041699349 |
| Name | traA_AVI_RS24150_pAtS4c |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123457.53 Da Isoelectric Point: 9.8997
>WP_041699349.1 Ti-type conjugative transfer relaxase TraA [Allorhizobium ampelinum]
MAIAHFSASIISRGDGRSVVLSAAYRHCAKMEYEREASTIDYTRKQGLLHEEFMLPADAPKWAKALIADR
SVSGAVEAFWNKVEAFEKRSDAQLARDLTIALPLELTPEQNIALVRDFVEKHILGKGMVADWVYHDNPGN
PHIHLMTTLRPLTEDGFGAKKVAVIGEDGQLVRTKSGKILYELWAGSTDDFNALRDGWFERLNHHLALGG
IDLRIDGRSYKKQAIELEPTIHLGVGAKAIARKAEQQGVRPELERIELNEERRSENTRRILKNPAIVLDL
IMREKSVFDERDVAKVLHRYVDDPAVFQQLMLRIILNPEVLRLQRDTIEFATGEKVPARYSTRAMIRLEA
TMARQAMWLSDKETHAVSTVVLAATFGRHGRLSEEQKAAIECIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIESRTLSSWELRWNRGRDVLDNKTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALALYN
ANARIVGERLKAEAVERLIADWNRGYDQTKTTLILAHLRRDVRMLNVMAREKLVERGIVGEGHVFRTADG
ERRYDAGDQIVFLKNETSLGVKNGMIGHVVEAEPNRIVAVVGDRDHRRHVVVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLAKVLSRKNAKETTLDYERGKLY
REALRFAENRGLHIMQVARTMLRDRLDWTLRQKTKVSDLVHRLRALGERLGLDQSPKTQTMKEAAPMVAG
IKTFSGSVADTVGDKLGADPTLKQQWEEVSVRFRYVFADPETAFRAINFDTVLADKEAAKAVLQKLEAEP
ASIGALKGKTGILASKTEREARRVAEVNVPALKRDLEQYLRMRETATLRIVTEEQALRQRVSVDIPALSP
AARVVLERVRDAIDRNDLPAALGYALSNRETKLEIDGFNQAVTERFGERTLLSNVAREPSGKLYEKLAEG
MKPEQKEQLKQAWPIMRTAQQLAAHERTVQSLKQAEELRQTLRQAPVLKQ
MAIAHFSASIISRGDGRSVVLSAAYRHCAKMEYEREASTIDYTRKQGLLHEEFMLPADAPKWAKALIADR
SVSGAVEAFWNKVEAFEKRSDAQLARDLTIALPLELTPEQNIALVRDFVEKHILGKGMVADWVYHDNPGN
PHIHLMTTLRPLTEDGFGAKKVAVIGEDGQLVRTKSGKILYELWAGSTDDFNALRDGWFERLNHHLALGG
IDLRIDGRSYKKQAIELEPTIHLGVGAKAIARKAEQQGVRPELERIELNEERRSENTRRILKNPAIVLDL
IMREKSVFDERDVAKVLHRYVDDPAVFQQLMLRIILNPEVLRLQRDTIEFATGEKVPARYSTRAMIRLEA
TMARQAMWLSDKETHAVSTVVLAATFGRHGRLSEEQKAAIECIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIESRTLSSWELRWNRGRDVLDNKTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALALYN
ANARIVGERLKAEAVERLIADWNRGYDQTKTTLILAHLRRDVRMLNVMAREKLVERGIVGEGHVFRTADG
ERRYDAGDQIVFLKNETSLGVKNGMIGHVVEAEPNRIVAVVGDRDHRRHVVVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLAKVLSRKNAKETTLDYERGKLY
REALRFAENRGLHIMQVARTMLRDRLDWTLRQKTKVSDLVHRLRALGERLGLDQSPKTQTMKEAAPMVAG
IKTFSGSVADTVGDKLGADPTLKQQWEEVSVRFRYVFADPETAFRAINFDTVLADKEAAKAVLQKLEAEP
ASIGALKGKTGILASKTEREARRVAEVNVPALKRDLEQYLRMRETATLRIVTEEQALRQRVSVDIPALSP
AARVVLERVRDAIDRNDLPAALGYALSNRETKLEIDGFNQAVTERFGERTLLSNVAREPSGKLYEKLAEG
MKPEQKEQLKQAWPIMRTAQQLAAHERTVQSLKQAEELRQTLRQAPVLKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16221 | GenBank | WP_012650583 |
| Name | traG_AVI_RS24135_pAtS4c |
UniProt ID | _ |
| Length | 656 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 656 a.a. Molecular weight: 70897.29 Da Isoelectric Point: 9.8632
>WP_012650583.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium/Agrobacterium group]
MTVNRLLLLILPAIIMLAAMFATSGMEQRLAAFGTSPQAKLMLGRAGLALPYIVAAAIGVIALFAANGSV
NVKAAGLSVLTGNGMVIIIAIARETIRLNGIAGSVPAGQSVLAYTDPVTMIGAGIAFISGMFALRVGIKG
NAAFETVAPKRIGGKRAVHGEADWMKIQEAAKLFPEPGGIVIGERYRVDRDSVAAMPFRADDRQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSNEVAPMVSEHRRQAGRKVIVLD
PTAGGVGFNALDWIGRHGNTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVCLSGHTEA
EDQTLRRVRANLSEPEPKLRARLTKIYEGSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
GLVSGVSFSTDDLADGGTDIFIALDLKVLEAHPGFARVVIGSLLNAIYNRNGNVKGRTLFLLDEVARLGY
LRILETARDAGRKYGITLTMIFQSLGQMREAYGGRDATSKWFESASWISFSAINDPDTADYISKRCGDTT
VEVDQTNRSTGMKGSSRSRSKQLSRRPLILPHEVLRMRADEQIVFTSGNPPLRCGRAIWFRREDMKACVG
GNRFHRTGTDTHTEHAQPRQKERKRP
MTVNRLLLLILPAIIMLAAMFATSGMEQRLAAFGTSPQAKLMLGRAGLALPYIVAAAIGVIALFAANGSV
NVKAAGLSVLTGNGMVIIIAIARETIRLNGIAGSVPAGQSVLAYTDPVTMIGAGIAFISGMFALRVGIKG
NAAFETVAPKRIGGKRAVHGEADWMKIQEAAKLFPEPGGIVIGERYRVDRDSVAAMPFRADDRQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSNEVAPMVSEHRRQAGRKVIVLD
PTAGGVGFNALDWIGRHGNTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVCLSGHTEA
EDQTLRRVRANLSEPEPKLRARLTKIYEGSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
GLVSGVSFSTDDLADGGTDIFIALDLKVLEAHPGFARVVIGSLLNAIYNRNGNVKGRTLFLLDEVARLGY
LRILETARDAGRKYGITLTMIFQSLGQMREAYGGRDATSKWFESASWISFSAINDPDTADYISKRCGDTT
VEVDQTNRSTGMKGSSRSRSKQLSRRPLILPHEVLRMRADEQIVFTSGNPPLRCGRAIWFRREDMKACVG
GNRFHRTGTDTHTEHAQPRQKERKRP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16222 | GenBank | WP_012650638 |
| Name | t4cp2_AVI_RS24425_pAtS4c |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 91946.71 Da Isoelectric Point: 5.7997
>WP_012650638.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium/Agrobacterium group]
MVALKLFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINMILARLG
SGWIIQVEALRVPTIDYPAGDACHFPDPVTRAIDSERRYHFERESGHFESRHALILTWQPPEPRRSGLTR
YVYSDTASRSARYADTTLDSFRTSIREVEQYLANVVSIRRMVTRETEERGAFRIARYDELFQFIRFCITG
ENHAVRLPDIPMYLDWLVTAQLQHGLTPTVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFVF
LDEQDARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAEAASQLVAYGYYTPVIVL
FDEQQPRLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLVNTRNLADLIPLNSVWS
GSPIAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHALVFGPTGSGKSTLLSLIAAQFRRYASAQIFA
FDKGRSMLPLTLAIGGDHYEIGGDTAGEGEGASPRLAFCPLAELSSDGDRAWAAEWIETLVALQGVTVTP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGAFQCFEVEEL
MNLGERNLVPVLTYMFRRVEKRLTGAPSLIILDEAWLMLGHPTFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDM
SLGPIALSFVGASGKEDLKRIRALHSKYGADWPKHWLQQRGIAHADTLFPAD
MVALKLFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINMILARLG
SGWIIQVEALRVPTIDYPAGDACHFPDPVTRAIDSERRYHFERESGHFESRHALILTWQPPEPRRSGLTR
YVYSDTASRSARYADTTLDSFRTSIREVEQYLANVVSIRRMVTRETEERGAFRIARYDELFQFIRFCITG
ENHAVRLPDIPMYLDWLVTAQLQHGLTPTVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFVF
LDEQDARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAEAASQLVAYGYYTPVIVL
FDEQQPRLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLVNTRNLADLIPLNSVWS
GSPIAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHALVFGPTGSGKSTLLSLIAAQFRRYASAQIFA
FDKGRSMLPLTLAIGGDHYEIGGDTAGEGEGASPRLAFCPLAELSSDGDRAWAAEWIETLVALQGVTVTP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGAFQCFEVEEL
MNLGERNLVPVLTYMFRRVEKRLTGAPSLIILDEAWLMLGHPTFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDM
SLGPIALSFVGASGKEDLKRIRALHSKYGADWPKHWLQQRGIAHADTLFPAD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 201057..210732
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AVI_RS24375 (Avi_9252) | 197317..198570 | + | 1254 | WP_012650628 | phosphonoacetate hydrolase | - |
| AVI_RS24380 (Avi_9253) | 198594..199517 | - | 924 | WP_012650629 | LysR substrate-binding domain-containing protein | - |
| AVI_RS29925 | 200236..200478 | + | 243 | WP_080517067 | PhnD/SsuA/transferrin family substrate-binding protein | - |
| AVI_RS29930 | 200459..200683 | + | 225 | WP_080517068 | hypothetical protein | - |
| AVI_RS30735 | 200764..201024 | + | 261 | WP_139192268 | ATP-binding cassette domain-containing protein | - |
| AVI_RS24390 (Avi_9257) | 201057..202370 | - | 1314 | WP_012650631 | IncP-type conjugal transfer protein TrbI | virB10 |
| AVI_RS24395 (Avi_9258) | 202382..202858 | - | 477 | WP_012650632 | conjugal transfer protein TrbH | - |
| AVI_RS24400 (Avi_9259) | 202858..203712 | - | 855 | WP_041699433 | P-type conjugative transfer protein TrbG | virB9 |
| AVI_RS24405 (Avi_9260) | 203730..204392 | - | 663 | WP_012650634 | conjugal transfer protein TrbF | virB8 |
| AVI_RS24410 (Avi_9261) | 204407..205603 | - | 1197 | WP_012650635 | P-type conjugative transfer protein TrbL | virB6 |
| AVI_RS24415 (Avi_9262) | 205597..205818 | - | 222 | WP_012650636 | entry exclusion protein TrbK | - |
| AVI_RS24420 (Avi_9263) | 205815..206624 | - | 810 | WP_012650637 | P-type conjugative transfer protein TrbJ | virB5 |
| AVI_RS24425 (Avi_9265) | 206596..209064 | - | 2469 | WP_012650638 | conjugal transfer protein TrbE | virb4 |
| AVI_RS24430 (Avi_9267) | 209075..209374 | - | 300 | WP_012650639 | conjugal transfer protein TrbD | virB3 |
| AVI_RS24435 (Avi_9268) | 209367..209771 | - | 405 | WP_012650640 | conjugal transfer pilin TrbC | virB2 |
| AVI_RS24440 (Avi_9269) | 209761..210732 | - | 972 | WP_012650641 | P-type conjugative transfer ATPase TrbB | virB11 |
| AVI_RS24445 (Avi_9270) | 210729..211364 | - | 636 | WP_041699366 | acyl-homoserine-lactone synthase | - |
Host bacterium
| ID | 22406 | GenBank | NC_011984 |
| Plasmid name | pAtS4c | Incompatibility group | - |
| Plasmid size | 211620 bp | Coordinate of oriT [Strand] | 145285..145324 [+] |
| Host baterium | Allorhizobium ampelinum S4 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixA, fixB |
| Anti-CRISPR | - |