Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121967 |
| Name | oriT_pAt2788a |
| Organism | Agrobacterium fabrum strain 2788 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP116685 (68812..68840 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pAt2788a
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file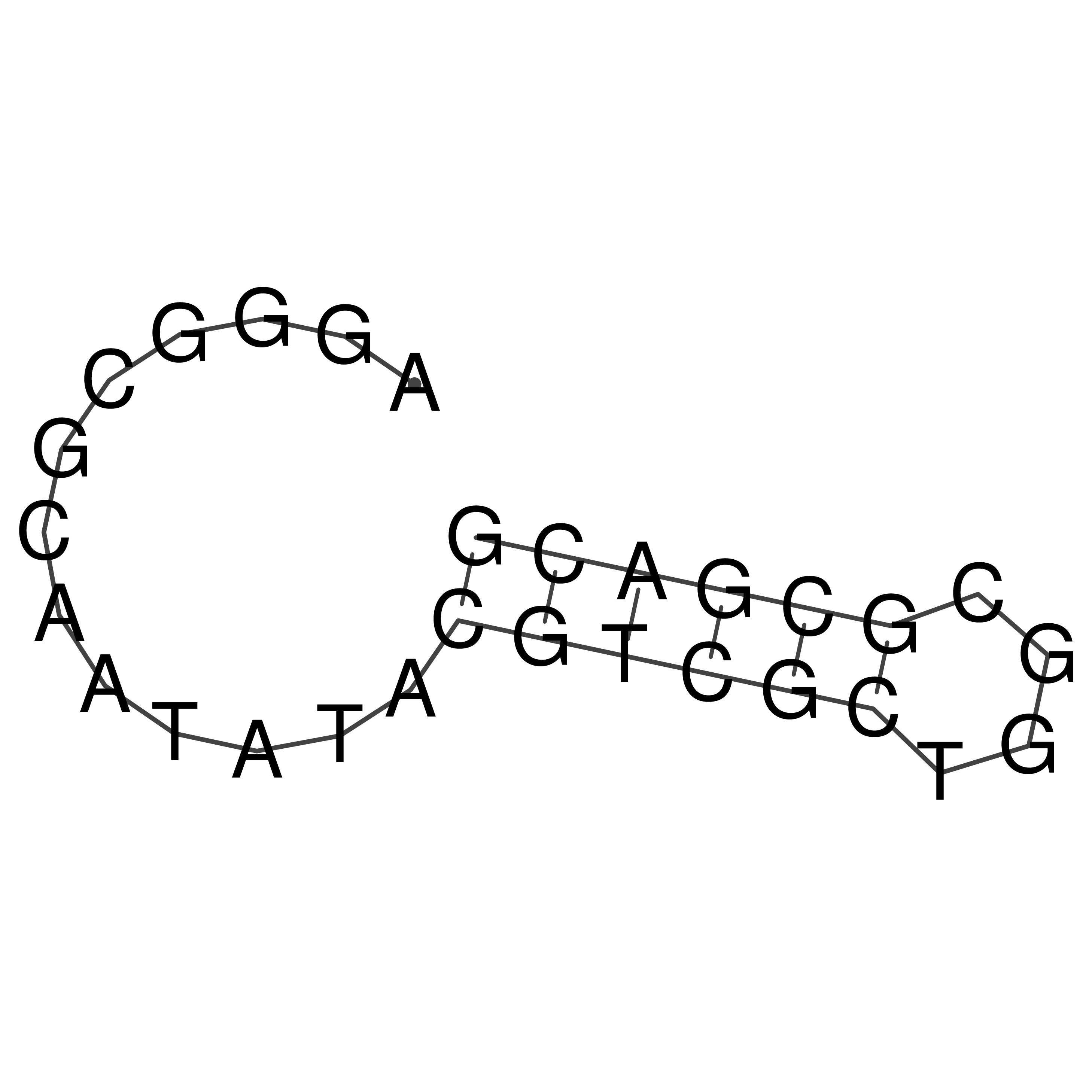
Relaxase
| ID | 13844 | GenBank | WP_173993503 |
| Name | traA_G6L39_RS23455_pAt2788a |
UniProt ID | _ |
| Length | 1542 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1542 a.a. Molecular weight: 171796.90 Da Isoelectric Point: 9.7206
>WP_173993503.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium fabrum]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTFFSYRGGAAELKHEELALPDQVPAWLRAAIDG
KSVVAASEAFWNAVDAFEARADAQLARELIIALPEELTRSENIALVREFVRDNLTSKGMIADWVYHDRQG
NPHIHLMTALRPLTEQGFGPKKVPVLGTDGEPLRVVTPDRPKGKIVYRLWAGDKETMKAWKIAWADMANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVAMYFAPADLARRQEMADRLLADPYLLLKQ
LGNERSTFDERDIAKALHRYVDDPADFANIRSKLTASNDLVLLKPQQANLQTGKVAEPAVFTTRDILRTE
YDMAQSAEVLARRKGFGVANAKIAAAVRTIETGNPENPFTLDREQVEAVHHVAGDTGIAAVVGHAGAGKS
TLLAAARVAWESDNYRVFGAALAGKAAEGLEDSSGIRSRTLASWEMAWEKGRDLLDRRDIFVIDEAGMVS
SQQMARVLKRAEEAGAKVVLVGDAMQLQPIQAGAAFRAISERIGSAELAGVRRQREEWAREASRLFARGK
VEEGLDAYAQRGRIVEAESRAEAVERIVADWTRARRDLIQQYADKEQPVRLRGDELLVLAHTNDDVRRLN
EALRNVMKDEGALTVSREFQTERGVREFAVGDRIIFLQNARFLEPRAQRLGPQYVKNGMLGTVVSTGDTH
GGPLLSVRLDNGRDVIVSEDSYRNVDHGYAATIHKSQGATVDRTFVLATGLMDQHLTYVSMTRHRDRADL
YAAREDFEPKPGWGKISRIDHAAGVTGELVGTGNAKFRPQDEDADKSPYADVKTDDGNVHRLWGVSLPKA
LEEAGVSEGDTVTLRKDGVERVTVMVPVIDEKTGKKRIEERTVHRNVWTAKRIETTEVRRQRLEEESHRP
ELFKQLVERLGRSGAKTTTLDFESEAGYRAHARDFAQRRGIDTLSGVAAWMEDGAARQLAWLAQKREQVA
KLWERASVALGFAIESEKRVSYSEERAETRTAKIPTDRHYLIPPTTLFSRSVGEDARRAQLRSDHWKERD
AILRPVLQKIYRDPEGALARLNTLASDARIAPRELADDLEASPQRLGRLHGSDLLVDGRAARSERDAAVS
ALSELLPLARAHATEFRRQVEHFGMREEQRRAHMSLSIPALSKQAMARLAEIEAVRERGGKDAYKTAFAY
AAEDRLLLREVKAVNDALNARFGWSAFTDKADTIAERKMIERMPDDLDLDRREKLNRLFAAIRRFAEEQH
LAERKDRSRIVTGASVVPGKETVPVLPMLAAVMEFETPIDHEARERARAVPHYSQNRAALVNAAARIWRD
PAGAAGKIEDLILKDFAGERIAAAISNDPAAYGALRGSGRIMDKLLAVGLERREAFRAVPEAGSRVRSLA
TSFRAAFDAEREAITEERQRMRIAIPGLSPSAEDALRELSVAMKKKNGRLDVAAGSLDPSISKEFATVSR
ALDERFGRNAILRGELDVINRVPPAQRRAFEVMREQLKVLQQVVRMQASQNIVEERHRRVLDRARGVILF
NQ
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTFFSYRGGAAELKHEELALPDQVPAWLRAAIDG
KSVVAASEAFWNAVDAFEARADAQLARELIIALPEELTRSENIALVREFVRDNLTSKGMIADWVYHDRQG
NPHIHLMTALRPLTEQGFGPKKVPVLGTDGEPLRVVTPDRPKGKIVYRLWAGDKETMKAWKIAWADMANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVAMYFAPADLARRQEMADRLLADPYLLLKQ
LGNERSTFDERDIAKALHRYVDDPADFANIRSKLTASNDLVLLKPQQANLQTGKVAEPAVFTTRDILRTE
YDMAQSAEVLARRKGFGVANAKIAAAVRTIETGNPENPFTLDREQVEAVHHVAGDTGIAAVVGHAGAGKS
TLLAAARVAWESDNYRVFGAALAGKAAEGLEDSSGIRSRTLASWEMAWEKGRDLLDRRDIFVIDEAGMVS
SQQMARVLKRAEEAGAKVVLVGDAMQLQPIQAGAAFRAISERIGSAELAGVRRQREEWAREASRLFARGK
VEEGLDAYAQRGRIVEAESRAEAVERIVADWTRARRDLIQQYADKEQPVRLRGDELLVLAHTNDDVRRLN
EALRNVMKDEGALTVSREFQTERGVREFAVGDRIIFLQNARFLEPRAQRLGPQYVKNGMLGTVVSTGDTH
GGPLLSVRLDNGRDVIVSEDSYRNVDHGYAATIHKSQGATVDRTFVLATGLMDQHLTYVSMTRHRDRADL
YAAREDFEPKPGWGKISRIDHAAGVTGELVGTGNAKFRPQDEDADKSPYADVKTDDGNVHRLWGVSLPKA
LEEAGVSEGDTVTLRKDGVERVTVMVPVIDEKTGKKRIEERTVHRNVWTAKRIETTEVRRQRLEEESHRP
ELFKQLVERLGRSGAKTTTLDFESEAGYRAHARDFAQRRGIDTLSGVAAWMEDGAARQLAWLAQKREQVA
KLWERASVALGFAIESEKRVSYSEERAETRTAKIPTDRHYLIPPTTLFSRSVGEDARRAQLRSDHWKERD
AILRPVLQKIYRDPEGALARLNTLASDARIAPRELADDLEASPQRLGRLHGSDLLVDGRAARSERDAAVS
ALSELLPLARAHATEFRRQVEHFGMREEQRRAHMSLSIPALSKQAMARLAEIEAVRERGGKDAYKTAFAY
AAEDRLLLREVKAVNDALNARFGWSAFTDKADTIAERKMIERMPDDLDLDRREKLNRLFAAIRRFAEEQH
LAERKDRSRIVTGASVVPGKETVPVLPMLAAVMEFETPIDHEARERARAVPHYSQNRAALVNAAARIWRD
PAGAAGKIEDLILKDFAGERIAAAISNDPAAYGALRGSGRIMDKLLAVGLERREAFRAVPEAGSRVRSLA
TSFRAAFDAEREAITEERQRMRIAIPGLSPSAEDALRELSVAMKKKNGRLDVAAGSLDPSISKEFATVSR
ALDERFGRNAILRGELDVINRVPPAQRRAFEVMREQLKVLQQVVRMQASQNIVEERHRRVLDRARGVILF
NQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16206 | GenBank | WP_173993502 |
| Name | traG_G6L39_RS23440_pAt2788a |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70592.61 Da Isoelectric Point: 9.1485
>WP_173993502.1 Ti-type conjugative transfer system protein TraG [Agrobacterium fabrum]
MKGKAKPRPSLLLIIVPIAVTGFSFYVAHWRWPELATGITGKTQYWFLRASPLPVLLFGPLAGLLAVWAL
PLHRRRPVALASFLCFLLMAGFYGMREFGRLQPLVESGVLTWDQALSFVDMIAVVGAAMGFMAAAVAARI
SSVVPEQVTRARRGTFGDADWLPMSAAARLFPDDGEIVIGERYRVDKELVHELPFDPKDPATWGRGGKAP
LLTYAQDFDSTHMLFFAGSGGFKTTSNVVPTALRYTGPLICLDPSTEVAPMVVDHRRDKLDREVVVLDPA
NPVMGFNVLDGIEHSLKKEEDIVGIAHMLLSESLRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQEHSASAFIRETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFKSSEIAGGKKDVFINIPASILRSYPGIGRVIIGSLINAMIEADGAFTRRALFILDEVDLLGYMRAL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNLGWGAKNSASRKSESITFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDMHAR
PNRFAKLGP
MKGKAKPRPSLLLIIVPIAVTGFSFYVAHWRWPELATGITGKTQYWFLRASPLPVLLFGPLAGLLAVWAL
PLHRRRPVALASFLCFLLMAGFYGMREFGRLQPLVESGVLTWDQALSFVDMIAVVGAAMGFMAAAVAARI
SSVVPEQVTRARRGTFGDADWLPMSAAARLFPDDGEIVIGERYRVDKELVHELPFDPKDPATWGRGGKAP
LLTYAQDFDSTHMLFFAGSGGFKTTSNVVPTALRYTGPLICLDPSTEVAPMVVDHRRDKLDREVVVLDPA
NPVMGFNVLDGIEHSLKKEEDIVGIAHMLLSESLRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQEHSASAFIRETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFKSSEIAGGKKDVFINIPASILRSYPGIGRVIIGSLINAMIEADGAFTRRALFILDEVDLLGYMRAL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNLGWGAKNSASRKSESITFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMDMHAR
PNRFAKLGP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 140961..150565
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| G6L39_RS23735 (G6L39_023735) | 136297..136902 | - | 606 | WP_111805088 | TetR/AcrR family transcriptional regulator | - |
| G6L39_RS23740 (G6L39_023740) | 136964..137833 | + | 870 | WP_111805086 | DMT family transporter | - |
| G6L39_RS23745 (G6L39_023745) | 137953..138342 | - | 390 | WP_236759236 | hypothetical protein | - |
| G6L39_RS23750 (G6L39_023750) | 138522..138833 | - | 312 | WP_162687912 | hypothetical protein | - |
| G6L39_RS23755 (G6L39_023755) | 139202..139606 | - | 405 | WP_111805084 | host attachment protein | - |
| G6L39_RS23760 (G6L39_023760) | 139964..140326 | - | 363 | WP_013637404 | transcriptional regulator | - |
| G6L39_RS23765 (G6L39_023765) | 140420..140959 | + | 540 | WP_013637405 | hypothetical protein | - |
| G6L39_RS23770 (G6L39_023770) | 140961..141650 | + | 690 | WP_111805080 | type IV secretion system protein VirB1 | virB1 |
| G6L39_RS23775 (G6L39_023775) | 141647..141946 | + | 300 | WP_019565723 | TrbC/VirB2 family protein | virB2 |
| G6L39_RS23780 (G6L39_023780) | 141949..142290 | + | 342 | WP_087747435 | type IV secretion system protein VirB3 | virB3 |
| G6L39_RS23785 (G6L39_023785) | 142283..144664 | + | 2382 | WP_080865446 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| G6L39_RS23790 (G6L39_023790) | 144664..145362 | + | 699 | WP_020811839 | P-type DNA transfer protein VirB5 | virB5 |
| G6L39_RS23795 (G6L39_023795) | 145359..145592 | + | 234 | WP_020811838 | EexN family lipoprotein | - |
| G6L39_RS23800 (G6L39_023800) | 145597..146532 | + | 936 | WP_111792071 | type IV secretion system protein | virB6 |
| G6L39_RS23805 (G6L39_023805) | 146568..146834 | + | 267 | WP_087747439 | hypothetical protein | - |
| G6L39_RS23810 (G6L39_023810) | 146838..147509 | + | 672 | WP_003517248 | virB8 family protein | virB8 |
| G6L39_RS23815 (G6L39_023815) | 147506..148360 | + | 855 | WP_162687911 | P-type conjugative transfer protein VirB9 | virB9 |
| G6L39_RS23820 (G6L39_023820) | 148374..149546 | + | 1173 | WP_111805078 | type IV secretion system protein VirB10 | virB10 |
| G6L39_RS23825 (G6L39_023825) | 149555..150565 | + | 1011 | WP_111805076 | P-type DNA transfer ATPase VirB11 | virB11 |
| G6L39_RS23830 (G6L39_023830) | 150952..151482 | + | 531 | WP_244644098 | LysR substrate-binding domain-containing protein | - |
| G6L39_RS23835 (G6L39_023835) | 151666..152277 | + | 612 | WP_111805074 | glutathione S-transferase family protein | - |
| G6L39_RS23840 (G6L39_023840) | 152338..154395 | + | 2058 | WP_111805072 | pyridoxamine 5'-phosphate oxidase family protein | - |
| G6L39_RS23845 (G6L39_023845) | 154519..155067 | + | 549 | WP_111805070 | carboxymuconolactone decarboxylase family protein | - |
Host bacterium
| ID | 22394 | GenBank | NZ_CP116685 |
| Plasmid name | pAt2788a | Incompatibility group | - |
| Plasmid size | 494181 bp | Coordinate of oriT [Strand] | 68812..68840 [+] |
| Host baterium | Agrobacterium fabrum strain 2788 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | PFL_4714 |
| Symbiosis gene | - |
| Anti-CRISPR | - |