Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121952 |
| Name | oriT_pK190-1-B |
| Organism | Enterococcus faecalis strain K190-1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP116573 (9452..9489 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 7..12, 16..21 (AGTAAC..GTTACT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pK190-1-B
ATACGAAGTAACGAAGTTACTGCGTATAAGTGCGCCCT
ATACGAAGTAACGAAGTTACTGCGTATAAGTGCGCCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file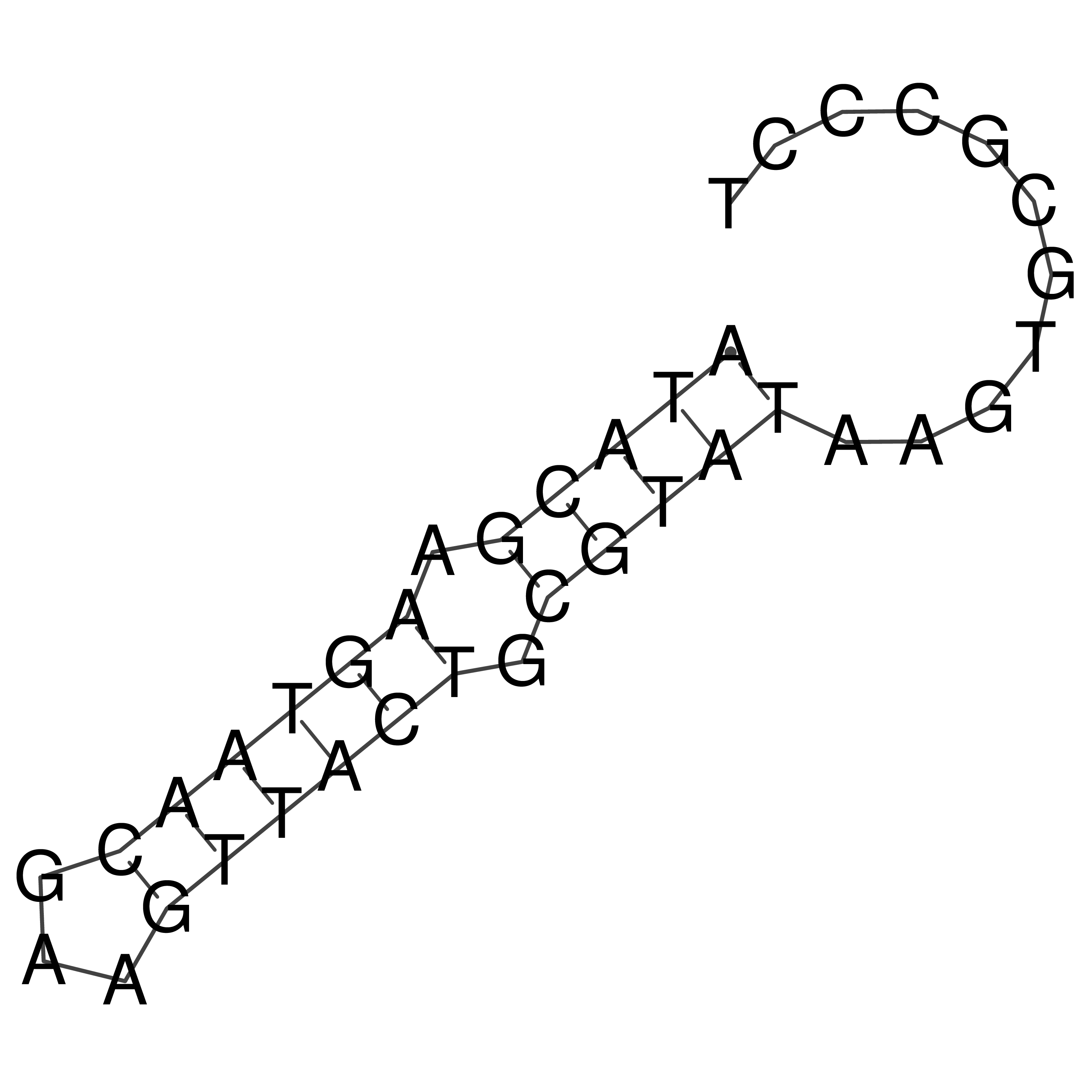
Relaxase
| ID | 13835 | GenBank | WP_002332652 |
| Name | mobQ_PML73_RS14825_pK190-1-B |
UniProt ID | _ |
| Length | 661 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 661 a.a. Molecular weight: 77293.34 Da Isoelectric Point: 9.3596
>WP_002332652.1 MULTISPECIES: MobQ family relaxase [Enterococcus]
MAIFHLSMTIAKRENGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTVKPEAFILKPDYVPNEFLDRQTL
WNKMELAEKSPNAQLCREVNVALPIELNNSDQRMLIEDFVKDNFVNEGMIADVAIHRDDENNPHAHIMLT
MREVDSEGNILNKSHRIPKLDENGNQIFNEKGQRVTVSIKTNDWGRKSLVSEIRKDWADKVNQYLKDRNI
DQQITEKSHAELGKKELPTIHEGFYSKKLEDKGVISELKRKNLEIQSYNDILAELDKLENQEKVLKQDQN
FTLKFEKTFSPLEKGELKNLSKELKLFINDENIDKRLGELKRWENSLIFNNKMEIQKQRLMLSKISSERD
MLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVVEKEESKK
IFKEKPFQTSRYLDSKIKQIEDSITKENNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVSIDS
VIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMKHID
ELSPLAKQNLKKVVNRNAYLPDSDKIELSKEIENTNKDQSQELDKDVPEKNEVTVKMFQFAKSINRLLSG
NQLQKKRNLDKLIKQTKAKKNQSLQRNIPLR
MAIFHLSMTIAKRENGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTVKPEAFILKPDYVPNEFLDRQTL
WNKMELAEKSPNAQLCREVNVALPIELNNSDQRMLIEDFVKDNFVNEGMIADVAIHRDDENNPHAHIMLT
MREVDSEGNILNKSHRIPKLDENGNQIFNEKGQRVTVSIKTNDWGRKSLVSEIRKDWADKVNQYLKDRNI
DQQITEKSHAELGKKELPTIHEGFYSKKLEDKGVISELKRKNLEIQSYNDILAELDKLENQEKVLKQDQN
FTLKFEKTFSPLEKGELKNLSKELKLFINDENIDKRLGELKRWENSLIFNNKMEIQKQRLMLSKISSERD
MLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVVEKEESKK
IFKEKPFQTSRYLDSKIKQIEDSITKENNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVSIDS
VIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMKHID
ELSPLAKQNLKKVVNRNAYLPDSDKIELSKEIENTNKDQSQELDKDVPEKNEVTVKMFQFAKSINRLLSG
NQLQKKRNLDKLIKQTKAKKNQSLQRNIPLR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16198 | GenBank | WP_032490379 |
| Name | t4cp2_PML73_RS14870_pK190-1-B |
UniProt ID | _ |
| Length | 551 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 551 a.a. Molecular weight: 63071.11 Da Isoelectric Point: 4.7399
>WP_032490379.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Lactobacillales]
MTSLLAESDGLILGKFPSGKVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFKTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLNLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVSEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKEYIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVTEEEEEKELSMSDILSSFDEGIEENTANNDNSELPF
MTSLLAESDGLILGKFPSGKVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFKTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLNLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVSEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKEYIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVTEEEEEKELSMSDILSSFDEGIEENTANNDNSELPF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 11947..25981
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PML73_RS14800 (PML73_14800) | 7027..7344 | + | 318 | WP_002326830 | hypothetical protein | - |
| PML73_RS14805 (PML73_14805) | 7878..8375 | + | 498 | WP_002338758 | DnaJ domain-containing protein | - |
| PML73_RS14810 (PML73_14810) | 8409..8642 | + | 234 | WP_002332654 | hypothetical protein | - |
| PML73_RS14815 (PML73_14815) | 8736..8993 | - | 258 | WP_000002668 | hypothetical protein | - |
| PML73_RS14820 (PML73_14820) | 8996..9295 | - | 300 | WP_002332653 | hypothetical protein | - |
| PML73_RS14825 (PML73_14825) | 9587..11572 | + | 1986 | WP_002332652 | MobQ family relaxase | - |
| PML73_RS14830 (PML73_14830) | 11596..11928 | + | 333 | WP_000713503 | CagC family type IV secretion system protein | - |
| PML73_RS14835 (PML73_14835) | 11947..12330 | + | 384 | WP_002332651 | hypothetical protein | trsC |
| PML73_RS14840 (PML73_14840) | 12347..12976 | + | 630 | WP_002332650 | hypothetical protein | trsD |
| PML73_RS14845 (PML73_14845) | 12987..14948 | + | 1962 | WP_002332649 | VirB4 family type IV secretion system protein | virb4 |
| PML73_RS14850 (PML73_14850) | 14962..16314 | + | 1353 | WP_002332648 | hypothetical protein | trsF |
| PML73_RS14855 (PML73_14855) | 16336..17445 | + | 1110 | WP_002332647 | lysozyme family protein | orf14 |
| PML73_RS14860 (PML73_14860) | 17458..18009 | + | 552 | WP_002332646 | hypothetical protein | - |
| PML73_RS14865 (PML73_14865) | 18014..18445 | + | 432 | WP_002332645 | hypothetical protein | - |
| PML73_RS14870 (PML73_14870) | 18438..20093 | + | 1656 | WP_032490379 | VirD4-like conjugal transfer protein, CD1115 family | - |
| PML73_RS14875 (PML73_14875) | 20111..21037 | + | 927 | WP_271865582 | hypothetical protein | traP |
| PML73_RS14880 (PML73_14880) | 21034..21966 | + | 933 | WP_104888282 | conjugal transfer protein TrbL family protein | trsL |
| PML73_RS14885 (PML73_14885) | 21983..22951 | + | 969 | WP_002377195 | hypothetical protein | - |
| PML73_RS14890 (PML73_14890) | 23000..23236 | + | 237 | Protein_29 | GNAT family N-acetyltransferase | - |
| PML73_RS14895 (PML73_14895) | 23237..24676 | + | 1440 | WP_001028144 | aminoglycoside O-phosphotransferase APH(2'')-Ia | - |
| PML73_RS14900 (PML73_14900) | 24768..25136 | + | 369 | WP_000516429 | hypothetical protein | - |
| PML73_RS14905 (PML73_14905) | 25196..25981 | + | 786 | WP_271865588 | LPXTG cell wall anchor domain-containing protein | prgC |
| PML73_RS14910 (PML73_14910) | 26287..27141 | - | 855 | WP_032490939 | aminoglycoside 6-adenylyltransferase | - |
| PML73_RS14915 (PML73_14915) | 27749..28297 | + | 549 | WP_250856472 | ATP-binding cassette domain-containing protein | - |
| PML73_RS14920 (PML73_14920) | 28598..28696 | - | 99 | WP_142435003 | type I toxin-antitoxin system Fst family toxin | - |
| PML73_RS14925 (PML73_14925) | 29119..29367 | + | 249 | WP_014862456 | helix-turn-helix domain-containing protein | - |
Host bacterium
| ID | 22379 | GenBank | NZ_CP116573 |
| Plasmid name | pK190-1-B | Incompatibility group | - |
| Plasmid size | 38206 bp | Coordinate of oriT [Strand] | 9452..9489 [-] |
| Host baterium | Enterococcus faecalis strain K190-1 |
Cargo genes
| Drug resistance gene | erm(B), aac(6')-aph(2'') |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |