Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 121843 |
| Name | oriT_pB |
| Organism | Sinorhizobium chiapasense strain S24 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP144207 (10398..10432 [+], 35 nt) |
| oriT length | 35 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 35 nt
>oriT_pB
TCCAAGGGCGCAATTATACGTCGCTGATGCGACGC
TCCAAGGGCGCAATTATACGTCGCTGATGCGACGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file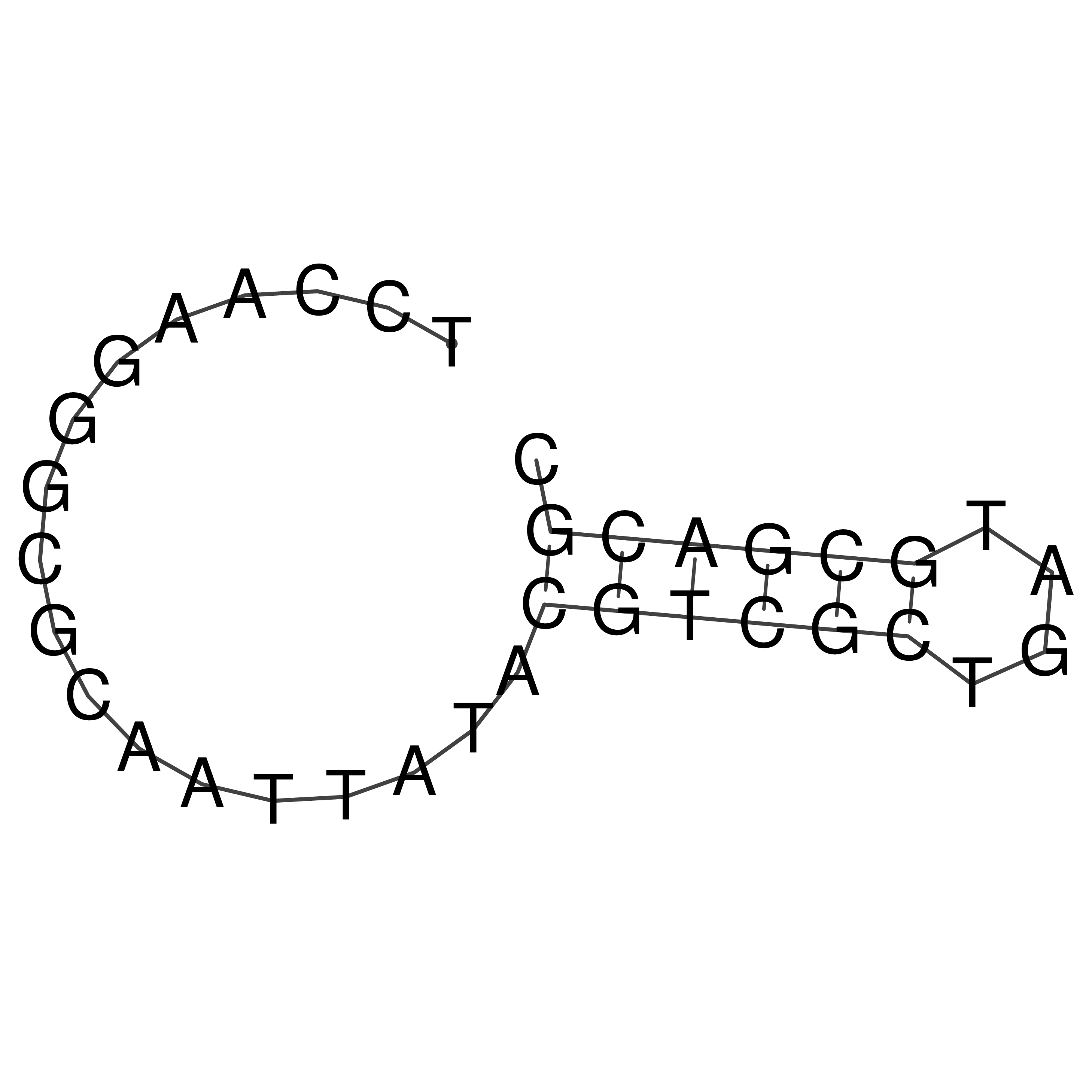
Relaxase
| ID | 13786 | GenBank | WP_334142256 |
| Name | traA_QM996_RS00080_pB |
UniProt ID | _ |
| Length | 1102 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1102 a.a. Molecular weight: 122936.89 Da Isoelectric Point: 9.8349
>WP_334142256.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium chiapasense]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVIPADSPEWLRSMIADR
SVASASEAFWNKVEDFEKRSDAQLAKDVTIALPIELTAEQNIALVRDFVERHVIAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGAKKIAVLGPDGNPLRNDAGKIVYELWAGSLDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKVTTEDQSVSLERLELQEEKRAENARRIQRRPEIVLDL
ITRENSVFDERDVAKILHRYIDDPGVFQSLMARILQSPETLRLERERIAFATGIRVPAKHTTRELIRLEA
EMGSRAIWLSRRSSHGVRSEVLEAAFSRHSRLSDEQKTAIEHVAGAERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGIASRTLSSWELRWNEGRNRLDDKTIFVLDEAGMVSSRQMALFVE
TATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDLARGNVGRAIDAYR
ANGRMMGSELKAEAVQNLVADWDRDYDPTTTTLILAHLRRDVRMLNEMARAKLVERGIVDAGFSFKTEDG
NRRFAPGDQIVFLKNEGALGVKNGMLGKVVEAAQNRIVAEIGEAGDRRQVTVESRFYNNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLGVYYGARSFTKSGGLIEILSRKDSKETTLDYEKATFY
RQALRFAEARGMHLVNVARTIARDRLQWTVRQKQKLNRLAYRLVAIGDKLGLAIGAKPINAQNLKETRPM
VAGIATFPKSIEQAVADKLEADPGLKKQWEDVSTRFHFVYAQPEAAFKAVNVDAILKDETVAKSTVAKIA
SRPDTYGALKGKTGLLASRADKRDRERAERNVPTLAQSLSDYMRQRSRAEQRYQAEELAVRRKVAIDIPA
LSPGARQTLERVRDAIDRNDLPAALEFALADKMVKAELEGFAKAVTERFGERTFLPLAAKDTNGEAFRTI
TAGMSPGQKAEVQTAWNTMRTVQQLNAHERTTEALRQAETLRQTKSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVIPADSPEWLRSMIADR
SVASASEAFWNKVEDFEKRSDAQLAKDVTIALPIELTAEQNIALVRDFVERHVIAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGAKKIAVLGPDGNPLRNDAGKIVYELWAGSLDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKVTTEDQSVSLERLELQEEKRAENARRIQRRPEIVLDL
ITRENSVFDERDVAKILHRYIDDPGVFQSLMARILQSPETLRLERERIAFATGIRVPAKHTTRELIRLEA
EMGSRAIWLSRRSSHGVRSEVLEAAFSRHSRLSDEQKTAIEHVAGAERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGIASRTLSSWELRWNEGRNRLDDKTIFVLDEAGMVSSRQMALFVE
TATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDLARGNVGRAIDAYR
ANGRMMGSELKAEAVQNLVADWDRDYDPTTTTLILAHLRRDVRMLNEMARAKLVERGIVDAGFSFKTEDG
NRRFAPGDQIVFLKNEGALGVKNGMLGKVVEAAQNRIVAEIGEAGDRRQVTVESRFYNNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLGVYYGARSFTKSGGLIEILSRKDSKETTLDYEKATFY
RQALRFAEARGMHLVNVARTIARDRLQWTVRQKQKLNRLAYRLVAIGDKLGLAIGAKPINAQNLKETRPM
VAGIATFPKSIEQAVADKLEADPGLKKQWEDVSTRFHFVYAQPEAAFKAVNVDAILKDETVAKSTVAKIA
SRPDTYGALKGKTGLLASRADKRDRERAERNVPTLAQSLSDYMRQRSRAEQRYQAEELAVRRKVAIDIPA
LSPGARQTLERVRDAIDRNDLPAALEFALADKMVKAELEGFAKAVTERFGERTFLPLAAKDTNGEAFRTI
TAGMSPGQKAEVQTAWNTMRTVQQLNAHERTTEALRQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 16120 | GenBank | WP_334141906 |
| Name | traG_QM996_RS00065_pB |
UniProt ID | _ |
| Length | 638 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 638 a.a. Molecular weight: 68814.60 Da Isoelectric Point: 8.6480
>WP_334141906.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium chiapasense]
MTANRITIAVAPLVLMVLAVIGMTGIEQWLSTFGKTEAARLILGRTGIAAPYITAASIGVIFLFASAGST
SIKFAGWGAAAGSIATILIAVMREATRLAAFAAQVPDGKSITSYLDHATMVGATAALMAGCFALRVALVG
NAAFARAEPKRIRGKRALHGEADWMKMQEAAKLFADTGGIVIGERYRVDKDSTAALLFRAGDPETWGAGG
KSSLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSNEVAPMVLEQRKGAARFIRILD
PKEPETGFNALDWIGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTEQ
EHQTLRQVRSNLAEPEPKLRERLQEIYDNSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYTNYA
ALVSGSTFSTDALADGKTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGEVKGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISKRCGMTT
VETDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRREDMAACVG
TNRFHPGG
MTANRITIAVAPLVLMVLAVIGMTGIEQWLSTFGKTEAARLILGRTGIAAPYITAASIGVIFLFASAGST
SIKFAGWGAAAGSIATILIAVMREATRLAAFAAQVPDGKSITSYLDHATMVGATAALMAGCFALRVALVG
NAAFARAEPKRIRGKRALHGEADWMKMQEAAKLFADTGGIVIGERYRVDKDSTAALLFRAGDPETWGAGG
KSSLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSNEVAPMVLEQRKGAARFIRILD
PKEPETGFNALDWIGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTEQ
EHQTLRQVRSNLAEPEPKLRERLQEIYDNSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYTNYA
ALVSGSTFSTDALADGKTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGEVKGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISKRCGMTT
VETDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRREDMAACVG
TNRFHPGG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 16121 | GenBank | WP_334141933 |
| Name | t4cp2_QM996_RS00155_pB |
UniProt ID | _ |
| Length | 817 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 817 a.a. Molecular weight: 91573.47 Da Isoelectric Point: 6.4279
>WP_334141933.1 conjugal transfer protein TrbE [Sinorhizobium chiapasense]
MVALKRFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
SGWMIQVEAVRIPTFDYPSEDRCHFPDPVTRAIDAERRAHFARERGHFESKHALILTHRPLESKKTALSK
YIYSDEESRKKTYADTVLFIFRNAIREIEQYLANTLSIRRMETREVVERGGERVTRYDELLQFVRFCITG
ENHPIRLPDAPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEAARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAQASSQLVTYGYYTPVIVL
FDGDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLVPLNSVWS
GNPIAPCPFYPANSPPLIQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYGQAQIFA
FDKGSSLLPLTLAARGDHYEIGGDAREAGAALAFCPLSDLENDADRAWATEWIEMLVALQGVTITPDHRN
AISRQVGLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLTLGAFQTFEIEHLMNMG
ERNLVPVLTYLFRRVEKRLDGSPSLILLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARELGTREFYERIGFNERQIEIVSTATPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSEHGRDWPIHWLQTRGVHDAATLLKA
MVALKRFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
SGWMIQVEAVRIPTFDYPSEDRCHFPDPVTRAIDAERRAHFARERGHFESKHALILTHRPLESKKTALSK
YIYSDEESRKKTYADTVLFIFRNAIREIEQYLANTLSIRRMETREVVERGGERVTRYDELLQFVRFCITG
ENHPIRLPDAPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEAARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAQASSQLVTYGYYTPVIVL
FDGDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLVPLNSVWS
GNPIAPCPFYPANSPPLIQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYGQAQIFA
FDKGSSLLPLTLAARGDHYEIGGDAREAGAALAFCPLSDLENDADRAWATEWIEMLVALQGVTITPDHRN
AISRQVGLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLTLGAFQTFEIEHLMNMG
ERNLVPVLTYLFRRVEKRLDGSPSLILLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARELGTREFYERIGFNERQIEIVSTATPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSEHGRDWPIHWLQTRGVHDAATLLKA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 15550..28996
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QM996_RS00085 (QM996_31170) | 13814..14380 | + | 567 | WP_334141911 | conjugative transfer signal peptidase TraF | - |
| QM996_RS00090 (QM996_31175) | 14370..15533 | + | 1164 | WP_334141913 | conjugal transfer protein TraB | - |
| QM996_RS00095 (QM996_31180) | 15550..16164 | + | 615 | WP_334141915 | TraH family protein | virB1 |
| QM996_RS00100 (QM996_31185) | 16250..17473 | - | 1224 | WP_334141917 | ATP-binding protein | - |
| QM996_RS00105 (QM996_31190) | 17667..17888 | - | 222 | WP_334141919 | helix-turn-helix transcriptional regulator | - |
| QM996_RS00110 (QM996_31195) | 18133..18453 | + | 321 | WP_334142258 | transcriptional repressor TraM | - |
| QM996_RS00115 (QM996_31200) | 18457..19167 | - | 711 | WP_334141921 | autoinducer binding domain-containing protein | - |
| QM996_RS00120 (QM996_31205) | 19490..20788 | - | 1299 | WP_334141923 | IncP-type conjugal transfer protein TrbI | virB10 |
| QM996_RS00125 (QM996_31210) | 20802..21239 | - | 438 | WP_334141924 | conjugal transfer protein TrbH | - |
| QM996_RS00130 (QM996_31215) | 21243..22055 | - | 813 | WP_334141926 | P-type conjugative transfer protein TrbG | virB9 |
| QM996_RS00135 (QM996_31220) | 22073..22735 | - | 663 | WP_334141928 | conjugal transfer protein TrbF | virB8 |
| QM996_RS00140 (QM996_31225) | 22759..23901 | - | 1143 | WP_334142260 | P-type conjugative transfer protein TrbL | virB6 |
| QM996_RS00145 (QM996_31230) | 23942..24118 | - | 177 | WP_334141929 | entry exclusion protein TrbK | - |
| QM996_RS00150 (QM996_31235) | 24112..24912 | - | 801 | WP_334141931 | P-type conjugative transfer protein TrbJ | virB5 |
| QM996_RS00155 (QM996_31240) | 24887..27340 | - | 2454 | WP_334141933 | conjugal transfer protein TrbE | virb4 |
| QM996_RS00160 (QM996_31245) | 27351..27650 | - | 300 | WP_334141934 | conjugal transfer protein TrbD | virB3 |
| QM996_RS00165 (QM996_31250) | 27643..28029 | - | 387 | WP_334141936 | TrbC/VirB2 family protein | virB2 |
| QM996_RS00170 (QM996_31255) | 28019..28996 | - | 978 | WP_334141938 | P-type conjugative transfer ATPase TrbB | virB11 |
| QM996_RS00175 (QM996_31260) | 29007..29633 | - | 627 | WP_334141940 | acyl-homoserine-lactone synthase | - |
| QM996_RS00180 (QM996_31265) | 29987..31210 | + | 1224 | WP_334142262 | plasmid partitioning protein RepA | - |
| QM996_RS00185 (QM996_31270) | 31207..32235 | + | 1029 | WP_334141942 | plasmid partitioning protein RepB | - |
| QM996_RS00190 (QM996_31275) | 32436..33647 | + | 1212 | WP_334141944 | plasmid replication protein RepC | - |
Host bacterium
| ID | 22270 | GenBank | NZ_CP144207 |
| Plasmid name | pB | Incompatibility group | - |
| Plasmid size | 444306 bp | Coordinate of oriT [Strand] | 10398..10432 [+] |
| Host baterium | Sinorhizobium chiapasense strain S24 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | bhpD, linJ, dxnH |
| Symbiosis gene | fixA, fixB |
| Anti-CRISPR | - |